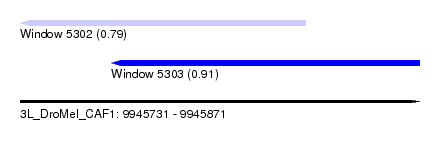
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,945,731 – 9,945,871 |
| Length | 140 |
| Max. P | 0.912190 |

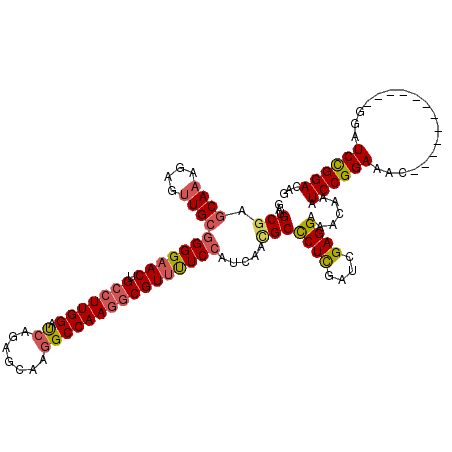
| Location | 9,945,731 – 9,945,831 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -30.11 |
| Energy contribution | -29.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
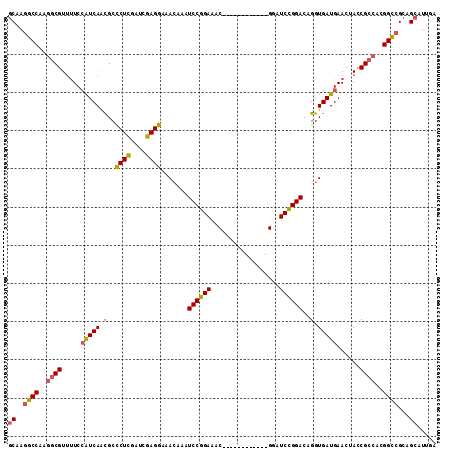
>3L_DroMel_CAF1 9945731 100 - 23771897 GCAAGGCCAAGGCGUUUUCCAUCAAUGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------------GAAUCCGGACAGGUGAUGAACUACCGCCAGGGUCGCAGCAUUGU ((..((((..((((.....(((((.((.((((....)))).......((((((...------------...))))))))..)))))......))))..))))...))..... ( -35.70) >DroSec_CAF1 97713 100 - 1 GCAAGGCCAAGGCGUUUUCCAUCAACGCUCUCGAUCGAGGAAACAAAUCCGGAAAC------------GGAUCCGGACAGGUGAUGAACUACCGCCAGGGUCGCAGCAUUGA ((..((((..((((.....(((((....(((.((((..(....).....((....)------------))))).)))....)))))......))))..))))...))..... ( -33.90) >DroSim_CAF1 101255 100 - 1 GCAAGGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------------GGAUCCGGACAGGUGAUGAACUACCGCCAGGGUCGCAGCAUUGA ((..((((..(((....(((........((((....))))......(((((....)------------))))..)))..((((......)))))))..))))...))..... ( -36.30) >DroEre_CAF1 100507 106 - 1 GCAAAGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------GUCUCCGGAUCCGGACAGGUGAUGAACUACCGCCACGGCCGCAGCAUUGA ((...(((..((((.....(((((.(..((((....)))).......((((((..(------(....))..))))))..).)))))......))))..))).))........ ( -34.80) >DroYak_CAF1 107531 106 - 1 GCAAGGCCAAGGCGUUUUCCAUCAACGCCCUUGAUCGAGGAAACAAAUCCGGAAAC------GUAUCCGGAUCCGGACAGGUGAUGAACUACCGCCACGGCCGCAGCAUUGA ((..((((..(((((((((((((((.....))))).).))))))...((((((..(------(....))..))))))..((((......)))))))..))))...))..... ( -39.00) >DroAna_CAF1 94510 112 - 1 CCAAGGCCAAAUCGUACUCGGUCAGCGCUCUCGAAAGAGGAAAUAAAUCCGGAAACAGGCCCGGAUCCGGAUCUGGACAGGUGAUGAACUAUCGUCACGGCCGCAGCAUUGA ....((((........((.(..(((...((((....))))......(((((((..(......)..))))))))))..)))(((((((....))))))))))).......... ( -38.20) >consensus GCAAGGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC____________GGAUCCGGACAGGUGAUGAACUACCGCCACGGCCGCAGCAUUGA ((..((((..((((.....(((((.(..((((....)))).......((((((..................))))))..).)))))......))))..))))...))..... (-30.11 = -29.92 + -0.19)

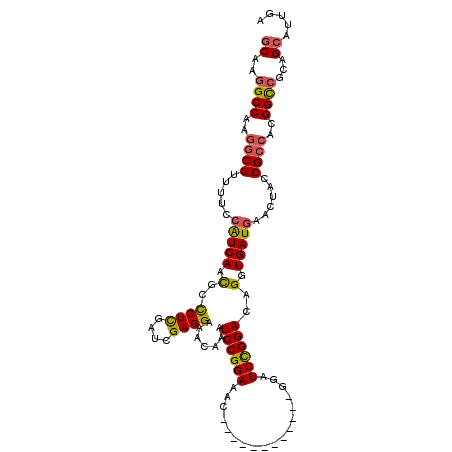
| Location | 9,945,763 – 9,945,871 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -34.42 |
| Energy contribution | -34.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9945763 108 - 23771897 GAUGCGAGCAAAGAGUUGCGGGGAACUGCCUUGGAUCAGAGCAAGGCCAAGGCGUUUUCCAUCAAUGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------------GAAUCCGGACAG ...(((.((((....))))((..(((.(((((((.((.......))))))))))))..)).....)))((((....)))).......((((((...------------...))))))... ( -40.60) >DroSec_CAF1 97745 108 - 1 GAUGCGAGCAAAGAGUUGCGGGGAACUGCCUUGGAUCAGAGCAAGGCCAAGGCGUUUUCCAUCAACGCUCUCGAUCGAGGAAACAAAUCCGGAAAC------------GGAUCCGGACAG (((.((((...((.((((.((..(((.(((((((.((.......))))))))))))..))..)))).)))))))))..(....)...((((((...------------...))))))... ( -42.30) >DroSim_CAF1 101287 108 - 1 GAUGCGAGCAAAGAGUUGCGGGGAACUGCCUUGGAUCAGAGCAAGGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------------GGAUCCGGACAG ...(((.((((....))))((..(((.(((((((.((.......))))))))))))..)).....)))((((....)))).......((((((...------------...))))))... ( -42.60) >DroEre_CAF1 100539 114 - 1 GAUGCGAGCAAGGAGUUGCGGGGAACUGCCUUGGAUCAGAGCAAAGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC------GUCUCCGGAUCCGGACAG (((.((((...((.((((.((..(((.(((((((............))))))))))..))..)))).)))))))))..(....)...((((((..(------(....))..))))))... ( -45.60) >DroYak_CAF1 107563 114 - 1 GAUGCGAGCAAGGAGCUGCGGGGAACUGCCUUGGAUCAGAGCAAGGCCAAGGCGUUUUCCAUCAACGCCCUUGAUCGAGGAAACAAAUCCGGAAAC------GUAUCCGGAUCCGGACAG ....(((.(((((.((((.((..(((.(((((((.((.......))))))))))))..))..))..))))))).))).(....)...((((((..(------(....))..))))))... ( -45.80) >DroAna_CAF1 94542 120 - 1 GAUGCCAGCAAAGAAUUGAAGGGCACGGCAUUGGACCAGACCAAGGCCAAAUCGUACUCGGUCAGCGCUCUCGAAAGAGGAAAUAAAUCCGGAAACAGGCCCGGAUCCGGAUCUGGACAG ((((((..(((....)))...)))..(((.((((......)))).)))..)))((..((((.......((((....))))........))))...(((..(((....)))..))).)).. ( -37.26) >consensus GAUGCGAGCAAAGAGUUGCGGGGAACUGCCUUGGAUCAGAGCAAGGCCAAGGCGUUUUCCAUCAACGCCCUCGAUCGAGGAAACAAAUCCGGAAAC____________GGAUCCGGACAG ...(((.((((....))))(((((((.(((((((.((.......)))))))))))))))).....)))((((....)))).......((((((..................))))))... (-34.42 = -34.87 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:57:01 2006