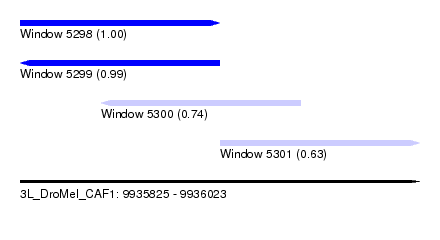
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,935,825 – 9,936,023 |
| Length | 198 |
| Max. P | 0.997769 |

| Location | 9,935,825 – 9,935,924 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.25 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -15.84 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9935825 99 + 23771897 GUGUCCGUCACUCAAUAUCACAUUCAGUCAAUAAUACGAUCUGAACGAAAUCGGCGAGGCUA---CGGCCAACGCCGAUUCCAGCAGAUA------CGGUUC------AGCC------AG (((((.((..............(((((((........)).))))).(.((((((((.(((..---..)))..)))))))).).)).))))------)((...------..))------.. ( -32.20) >DroVir_CAF1 88248 85 + 1 GUGUCCGUCACUCAAUAUAACAUUCAGUCAAUAAUAUAACCUGAACGAAAUCGGCGGGGCCC---AACCUCCGGCUGAUUACAACAAU-------------------------------- (((.....)))...........(((((.............))))).(.(((((((((((...---...)))).))))))).)......-------------------------------- ( -16.42) >DroPse_CAF1 88108 105 + 1 GUGUCCGUCACUCAAUAUCACAUUCAGUCAAUAAUAUAACCUGAACGAAAUCGGCGGGGCCC---AGCCCCCCGCCGAUUCGCUCAGAUU------CGGGUA------CGUCAAUCCAAG ((..(((..(((.(((.....))).)))............((((.((((.(((((((((...---....))))))))))))).))))...------)))..)------)........... ( -35.60) >DroWil_CAF1 133471 102 + 1 GUGUCCGUCACUCAAUAUAACAUUCAGUCAAUAGUAUAAUCUGAACGAAAUCGGUGAAUCGAGGCAAUCGAUCACCGAUUCGAUUAACAA------CAGCAG------GGAC------AG .(((((................(((((.............)))))((((.(((((((.((((.....)))))))))))))))........------......------))))------). ( -29.52) >DroAna_CAF1 85105 111 + 1 GUGUCCGUCACUCAAUAUUACAUUCAGUCAAUAGUACGAUCUGAACGAAAUCGGCGGGGCUC---CGGCCAUCGCCGAUUCCAGCAGAUAUUCGUACCGAUUGGCCCAAUCC------AG (((.....)))...............((((((.((((((((((...(.((((((((((((..---..))).))))))))).)..))))...))))))..)))))).......------.. ( -37.30) >DroPer_CAF1 89220 105 + 1 GUGUCCGUCACUCAAUAUCACAUUCAGUCAAUAAUAUAACCUGAACGAAAUCGGCGGGGCCC---AGCCCCCCGCCGAUUCGCUCAGAUU------CGGGUA------CGUCAAUCCAAG ((..(((..(((.(((.....))).)))............((((.((((.(((((((((...---....))))))))))))).))))...------)))..)------)........... ( -35.60) >consensus GUGUCCGUCACUCAAUAUCACAUUCAGUCAAUAAUAUAACCUGAACGAAAUCGGCGGGGCCC___AGCCCACCGCCGAUUCCAUCAGAUA______CGGGUA______AGCC______AG (((.....))).........(.(((((.............))))).).(((((((((((.........)).)))))))))........................................ (-15.84 = -15.70 + -0.13)


| Location | 9,935,825 – 9,935,924 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.25 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9935825 99 - 23771897 CU------GGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCG---UAGCCUCGCCGAUUUCGUUCAGAUCGUAUUAUUGACUGAAUGUGAUAUUGAGUGACGGACAC ..------....------...(((------(.((....((((((((((..(((..---..))).))))))))))((((((.(((......)))))))))........))...)))).... ( -35.20) >DroVir_CAF1 88248 85 - 1 --------------------------------AUUGUUGUAAUCAGCCGGAGGUU---GGGCCCCGCCGAUUUCGUUCAGGUUAUAUUAUUGACUGAAUGUUAUAUUGAGUGACGGACAC --------------------------------...((((....))))..((((((---((......))))))))((((.(((((......)))))....(((((.....))))))))).. ( -20.20) >DroPse_CAF1 88108 105 - 1 CUUGGAUUGACG------UACCCG------AAUCUGAGCGAAUCGGCGGGGGGCU---GGGCCCCGCCGAUUUCGUUCAGGUUAUAUUAUUGACUGAAUGUGAUAUUGAGUGACGGACAC ............------...(((------(((((((((((((((((((((....---...))))))))).)))))))))))).........(((.((((...)))).)))..))).... ( -42.60) >DroWil_CAF1 133471 102 - 1 CU------GUCC------CUGCUG------UUGUUAAUCGAAUCGGUGAUCGAUUGCCUCGAUUCACCGAUUUCGUUCAGAUUAUACUAUUGACUGAAUGUUAUAUUGAGUGACGGACAC .(------((((------(....)------..((((.((((((((((((((((.....)))).))))))))))(((((((.(((......)))))))))).......)).))))))))). ( -32.50) >DroAna_CAF1 85105 111 - 1 CU------GGAUUGGGCCAAUCGGUACGAAUAUCUGCUGGAAUCGGCGAUGGCCG---GAGCCCCGCCGAUUUCGUUCAGAUCGUACUAUUGACUGAAUGUAAUAUUGAGUGACGGACAC .(------(.(((.((.((((.(((((((...((((.(((((((((((..(((..---..))).)))))))))))..))))))))))))))).)).))).)).................. ( -39.20) >DroPer_CAF1 89220 105 - 1 CUUGGAUUGACG------UACCCG------AAUCUGAGCGAAUCGGCGGGGGGCU---GGGCCCCGCCGAUUUCGUUCAGGUUAUAUUAUUGACUGAAUGUGAUAUUGAGUGACGGACAC ............------...(((------(((((((((((((((((((((....---...))))))))).)))))))))))).........(((.((((...)))).)))..))).... ( -42.60) >consensus CU______GACG______UACCCG______UAUCUGAUCGAAUCGGCGGGGGGCU___GGGCCCCGCCGAUUUCGUUCAGAUUAUAUUAUUGACUGAAUGUGAUAUUGAGUGACGGACAC ................................((((...((((((((((..............))))))))))(((((((.(((......)))))))))).............))))... (-19.90 = -20.12 + 0.23)

| Location | 9,935,865 – 9,935,964 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -29.63 |
| Energy contribution | -30.49 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
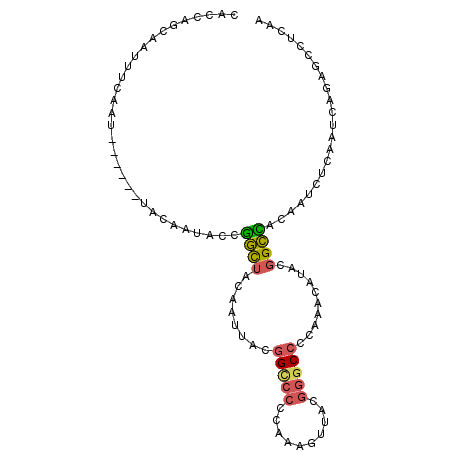
>3L_DroMel_CAF1 9935865 99 - 23771897 AUUGUAGCCGGUAUUGUAGCCGGUAUUGAAAUUGCUGGUGCUGGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG ...((((.((((....(((((((((((.........))))))))))------).))))------...)))).((((((((((..(((....))).))))))))))...... ( -42.70) >DroSec_CAF1 87864 93 - 1 AUUGUAGCCGGUAUUGUA------AUUGAAAUUGCUGGUGCUGGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG .....(((((((((((((------((....))))).))))))))))------((((.(------(....)).((((((((((..(((....))).)))))))))))))).. ( -39.80) >DroSim_CAF1 91423 93 - 1 AUUGUAGCCGGUAUUGUA------AUUGAAAUUGCUGGUGCUGGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG .....(((((((((((((------((....))))).))))))))))------((((.(------(....)).((((((((((..(((....))).)))))))))))))).. ( -39.80) >DroEre_CAF1 90484 93 - 1 AUUGUAGCCGGGAUUGUA------AUUGAAAUUGCUGGUGCUGGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG .....((((((.((((((------((....))))).))).))))))------((((.(------(....)).((((((((((..(((....))).)))))))))))))).. ( -37.30) >DroYak_CAF1 91946 93 - 1 AUUGUAGCCGGUAUUGUA------AUUGAAAUUGCUGGUGCUGGCU------GAACCG------UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG .....(((((((((((((------((....))))).))))))))))------((((.(------(....)).((((((((((..(((....))).)))))))))))))).. ( -39.80) >DroAna_CAF1 85145 105 - 1 AUUGUAGCCGGAUUGGUA------GCCGGAAUUGUAGCUGCUGGAUUGGGCCAAUCGGUACGAAUAUCUGCUGGAAUCGGCGAUGGCCGGAGCCCCGCCGAUUUCGUUCAG .(((((.(((.((((((.------.((((...........)))).....))))))))))))))....(((.(((((((((((..(((....))).)))))))))))..))) ( -40.00) >consensus AUUGUAGCCGGUAUUGUA______AUUGAAAUUGCUGGUGCUGGCU______GAACCG______UAUCUGCUGGAAUCGGCGUUGGCCGUAGCCUCGCCGAUUUCGUUCAG .....(((((((((((((..............))).)))))))))).....................(((.(((((((((((..(((....))).)))))))))))..))) (-29.63 = -30.49 + 0.86)
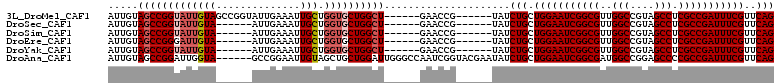
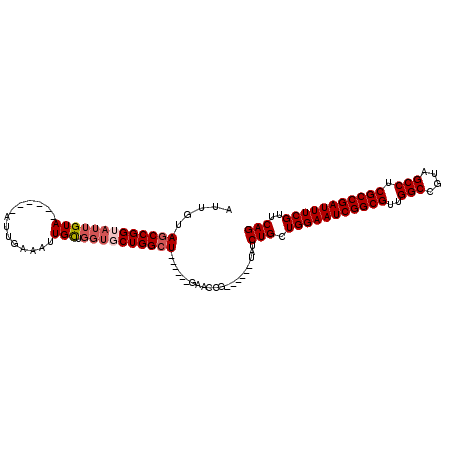
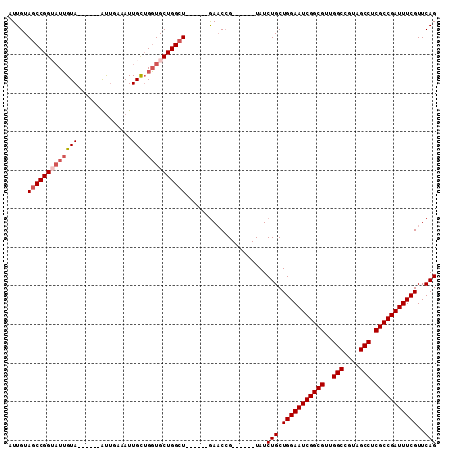
| Location | 9,935,924 – 9,936,023 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -8.75 |
| Energy contribution | -8.67 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9935924 99 + 23771897 CACCAGCAAUUUCAAUACCGGCUACAAUACCGGCUACAAUUACGGGCCCCAAAGUUACGGGCCACAAACAUACGGCCACAAUCUCAAUCAGAGCCUCAA ...................(((.........((((.........(((((.........)))))..........))))....(((.....)))))).... ( -19.91) >DroSec_CAF1 87923 93 + 1 CACCAGCAAUUUCAAU------UACAAUACCGGCUACAAUUACGGCCCCCAAAGUUACGGGCCCCAAACAUACGGCCACAAUCUCAAUCAGAGCCUCAA ................------.........((((........(((((..........)))))((........))................)))).... ( -15.00) >DroSim_CAF1 91482 93 + 1 CACCAGCAAUUUCAAU------UACAAUACCGGCUACAAUUACGGCCCCCAAAGUUACGGGCCCCAAACAUACGGCCACAAUCUCAAUCAGAGCCUCAA ................------.........((((........(((((..........)))))((........))................)))).... ( -15.00) >DroEre_CAF1 90543 93 + 1 CACCAGCAAUUUCAAU------UACAAUCCCGGCUACAAUUACGGCCCCCAAAGUUACGGGCCCCAAAGUUACGGCCACAAUCUCAAUCAGAGUCUCAA ................------.........((((..((((..(((((..........)))))....))))..)))).....(((.....)))...... ( -16.00) >DroWil_CAF1 133573 87 + 1 ACACAGAAAUAGCUAU------CACAAUAUAUAUGGCAGUAACUGUG-CCAAUGCCAGUGGC-----AGUGGCAGUGGCAAUUACAAUUACAAUUACAA ...........(((((------..........))))).((((...((-(((.(((((.....-----..))))).))))).)))).............. ( -20.70) >DroYak_CAF1 92005 93 + 1 CACCAGCAAUUUCAAU------UACAAUACCGGCUACAAUUACGGCCCCCAAAGUUACGGGCCCCAAAGCUAUGGCUACAAUCUCAAUCAGAGCCUCAA ................------.........((((........(((((..........)))))....(((....)))..............)))).... ( -17.30) >consensus CACCAGCAAUUUCAAU______UACAAUACCGGCUACAAUUACGGCCCCCAAAGUUACGGGCCCCAAACAUACGGCCACAAUCUCAAUCAGAGCCUCAA ...............................((((........(((((..........)))))..........))))...................... ( -8.75 = -8.67 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:59 2006