
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,893,668 – 9,893,758 |
| Length | 90 |
| Max. P | 0.974543 |

| Location | 9,893,668 – 9,893,758 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.40 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -11.10 |
| Energy contribution | -10.93 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
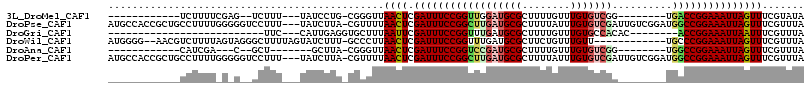
>3L_DroMel_CAF1 9893668 90 + 23771897 UAUACGAAACUAAUUUCCGGUCA--------CCGACACAAACAAAAGCGCAUCCAACCGGAAAUCGAGUUAACCCG-CAGGAUA---AAAGA--CUCGAAAAGA------------ .............((((((((..--------....(.(........).)......))))))))(((((((..((..-..))...---...))--))))).....------------ ( -15.80) >DroPse_CAF1 52796 112 + 1 UAAACGAAACUAAUUUCCGGCCAUCCGACAAUCGACACAAAUAAAAGCGCAUCAAGCCGGAAAUCGAGUUAAAACG-UAAGAUA---AAAGGACCCCCAAAAGGCAGCGGUGGCAU ...(((.((((.(((((((((....((.....)).(.(........).)......)))))))))..))))....))-)......---....(.(((((........).)).))).. ( -17.60) >DroGri_CAF1 54609 79 + 1 UAAACGAAAUUAAUUUCCGGU--------GUGUGGCACAAACAAAAGCGCAUCAAACCGGAAAUCGAAUUAAAGCACCUCAAUG---GAA-------------------------- .........(..(((((((((--------.((..((.(........).))..)).)))))))))..).........((.....)---)..-------------------------- ( -18.10) >DroWil_CAF1 85099 101 + 1 UAAACGAAACUAAUUUCCGGGCA------------AACAAACAGAAGCGCAUCAAACCGGAAAUCGAGUUAAGGGC-AAAGAUACUAAAAGCCCUACUAAAAGACGUU--CCCCAU ..((((.((((.((((((((...------------.....................))))))))..)))).(((((-.............))))).........))))--...... ( -21.58) >DroAna_CAF1 54382 83 + 1 UAAACGAAACUAAUUUCCGGCCA--------CCGACACAAACAAAAGCGCAUCGGACCGGAAAUCGAGUUAACCCG-UAAGC-------AGC--G---UCGAUG------------ ....(((((((.((((((((...--------((((..(..........)..)))).))))))))..))))....((-(....-------.))--)---)))...------------ ( -18.70) >DroPer_CAF1 53856 112 + 1 UAAACGAAACUAAUUUCCGGCCAUCCGACAAUCGACACAAAUAAAAGCGCAUCAAGCCGGAAAUCGAGUUAAAACG-UAAGAUA---AAAGGACCCCCAAAAGGCAGCGGUGGCAU ...(((.((((.(((((((((....((.....)).(.(........).)......)))))))))..))))....))-)......---....(.(((((........).)).))).. ( -17.60) >consensus UAAACGAAACUAAUUUCCGGCCA________CCGACACAAACAAAAGCGCAUCAAACCGGAAAUCGAGUUAAAACG_UAAGAUA___AAAGC__C_C_AAAAGG____________ .......((((.(((((((((..................................)))))))))..)))).............................................. (-11.10 = -10.93 + -0.17)


| Location | 9,893,668 – 9,893,758 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.40 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
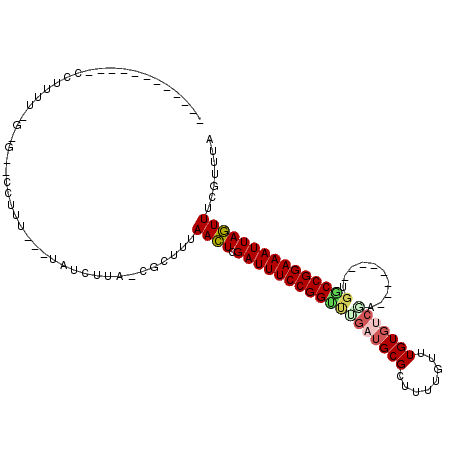
>3L_DroMel_CAF1 9893668 90 - 23771897 ------------UCUUUUCGAG--UCUUU---UAUCCUG-CGGGUUAACUCGAUUUCCGGUUGGAUGCGCUUUUGUUUGUGUCGG--------UGACCGGAAAUUAGUUUCGUAUA ------------.....(((((--(....---.((((..-.))))..))))))((((((((((...((((........))))...--------))))))))))............. ( -24.10) >DroPse_CAF1 52796 112 - 1 AUGCCACCGCUGCCUUUUGGGGGUCCUUU---UAUCUUA-CGUUUUAACUCGAUUUCCGGCUUGAUGCGCUUUUAUUUGUGUCGAUUGUCGGAUGGCCGGAAAUUAGUUUCGUUUA ........(((.((....)).))).....---......(-((....((((.(((((((((((((((((((........)))).....))))...))))))))))))))).)))... ( -29.30) >DroGri_CAF1 54609 79 - 1 --------------------------UUC---CAUUGAGGUGCUUUAAUUCGAUUUCCGGUUUGAUGCGCUUUUGUUUGUGCCACAC--------ACCGGAAAUUAAUUUCGUUUA --------------------------...---.((((((....))))))..((((((((((.((..((((........))))..)).--------))))))))))........... ( -20.70) >DroWil_CAF1 85099 101 - 1 AUGGGG--AACGUCUUUUAGUAGGGCUUUUAGUAUCUUU-GCCCUUAACUCGAUUUCCGGUUUGAUGCGCUUCUGUUUGUU------------UGCCCGGAAAUUAGUUUCGUUUA ....((--(((.......((((((((.............-)))))..))).(((((((((...((((.((....)).))))------------...))))))))).)))))..... ( -26.22) >DroAna_CAF1 54382 83 - 1 ------------CAUCGA---C--GCU-------GCUUA-CGGGUUAACUCGAUUUCCGGUCCGAUGCGCUUUUGUUUGUGUCGG--------UGGCCGGAAAUUAGUUUCGUUUA ------------...(((---.--(((-------(....-((((....))))(((((((((((((((((........))))))))--------..))))))))))))).))).... ( -30.10) >DroPer_CAF1 53856 112 - 1 AUGCCACCGCUGCCUUUUGGGGGUCCUUU---UAUCUUA-CGUUUUAACUCGAUUUCCGGCUUGAUGCGCUUUUAUUUGUGUCGAUUGUCGGAUGGCCGGAAAUUAGUUUCGUUUA ........(((.((....)).))).....---......(-((....((((.(((((((((((((((((((........)))).....))))...))))))))))))))).)))... ( -29.30) >consensus ____________CCUUUU_G_G__CCUUU___UAUCUUA_CGCUUUAACUCGAUUUCCGGUUUGAUGCGCUUUUGUUUGUGUCGA________UGGCCGGAAAUUAGUUUCGUUUA ..............................................((((.((((((((((((((((((........)))))))..........)))))))))))))))....... (-16.69 = -17.13 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:50 2006