
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
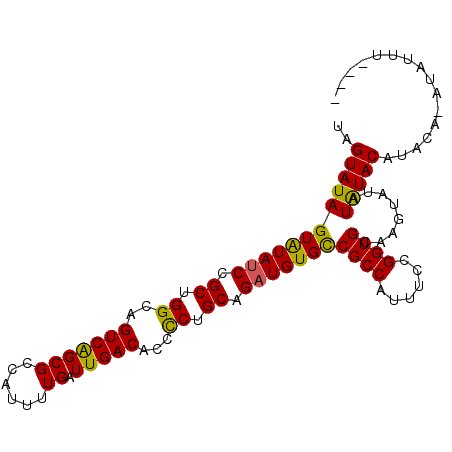
| Location | 9,889,068 – 9,889,165 |
| Length | 97 |
| Max. P | 0.845131 |

| Location | 9,889,068 – 9,889,165 |
|---|---|
| Length | 97 |
| Sequences | 4 |
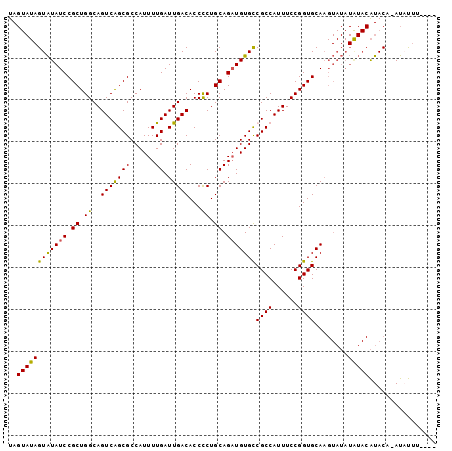
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -17.77 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9889068 97 + 23771897 UAGUAUAGUAUAUCCGCUGGCAGUCAGCGCCAUUUUGAUUGACACCCCUGCAGAUGUGCCGCCAUUUCCGGUGCAAGUAUAUGUACAUACA-AUAUUU---- ..((((((((((((.((.((..(((((((......)).)))))...)).)).)))))))((((......))))........))))).....-......---- ( -24.00) >DroSec_CAF1 49774 97 + 1 UAGUAUAGUAUAUCCGCUGGCAGUCGGCGCCAUUUUGAUUGACACCCCUGCAGAUGUGCCGCCAUUUCCGGUGCAAGUAUAUAUACAUACC-AUUUUU---- ..(((((.(((((.((((((.(((.((((.(((((((...(......)..)))).))).))))))).))))))...)))))))))).....-......---- ( -25.30) >DroSim_CAF1 51886 96 + 1 UAGUAUAGUAUAUCCGCUGGCAGUCGGCGCCAUUUUGAUUGACACCCCUGCAGUUGUGCCGCCAUUUCCGGUGCAAGUAUAUAUACAUACC-AUUUU----- ..(((((.(((((.((((((.(((.((((.(((...(((((.((....)))))))))).))))))).))))))...)))))))))).....-.....----- ( -26.40) >DroEre_CAF1 52097 86 + 1 UA-----GUGUAUCCGCUGGCAGUCAGCGCCAUUUUGAUUGACACCUCUGCAGAUGUGUCGCCAUUUCCGGUGCA-----------GUACACACACAUAUAU ..-----(((((((((.((((((((((.......))))))(((((.((....)).)))))))))....)))....-----------)))))).......... ( -24.80) >consensus UAGUAUAGUAUAUCCGCUGGCAGUCAGCGCCAUUUUGAUUGACACCCCUGCAGAUGUGCCGCCAUUUCCGGUGCAAGUAUAUAUACAUACA_AUAUUU____ ..((((((((((((.((.((..(((((((......)).)))))...)).)).)))))))((((......))))........)))))................ (-17.77 = -17.40 + -0.37)

| Location | 9,889,068 – 9,889,165 |
|---|---|
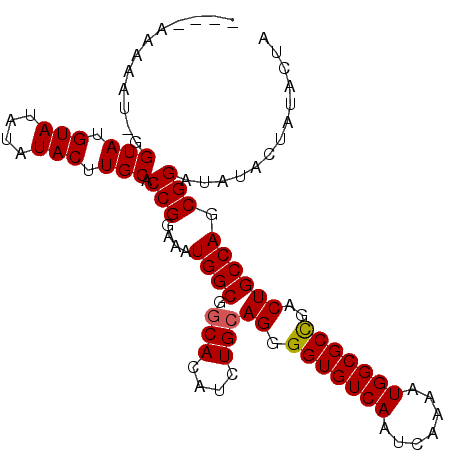
| Length | 97 |
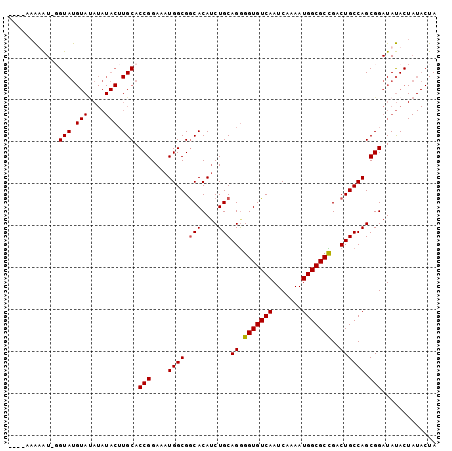
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9889068 97 - 23771897 ----AAAUAU-UGUAUGUACAUAUACUUGCACCGGAAAUGGCGGCACAUCUGCAGGGGUGUCAAUCAAAAUGGCGCUGACUGCCAGCGGAUAUACUAUACUA ----...(((-.((((((.((......))..(((....((((.(((....)))((.(((((((.......)))))))..)))))).))))))))).)))... ( -22.90) >DroSec_CAF1 49774 97 - 1 ----AAAAAU-GGUAUGUAUAUAUACUUGCACCGGAAAUGGCGGCACAUCUGCAGGGGUGUCAAUCAAAAUGGCGCCGACUGCCAGCGGAUAUACUAUACUA ----.....(-(((((((((((.....(((.(((....)))..)))...((((((.(((((((.......)))))))..))).)))...)))))).)))))) ( -28.30) >DroSim_CAF1 51886 96 - 1 -----AAAAU-GGUAUGUAUAUAUACUUGCACCGGAAAUGGCGGCACAACUGCAGGGGUGUCAAUCAAAAUGGCGCCGACUGCCAGCGGAUAUACUAUACUA -----....(-(((((((((((.....(((.(((....)))..)))(..((((((.(((((((.......)))))))..))).)))..))))))).)))))) ( -30.10) >DroEre_CAF1 52097 86 - 1 AUAUAUGUGUGUGUAC-----------UGCACCGGAAAUGGCGACACAUCUGCAGAGGUGUCAAUCAAAAUGGCGCUGACUGCCAGCGGAUACAC-----UA ........((((((.(-----------(((...(((..((....))..)))((((.(((((((.......)))))))..))))..))))))))))-----.. ( -28.90) >consensus ____AAAAAU_GGUAUGUAUAUAUACUUGCACCGGAAAUGGCGGCACAUCUGCAGGGGUGUCAAUCAAAAUGGCGCCGACUGCCAGCGGAUAUACUAUACUA ............(((.(((....))).))).(((....((((.(((....)))((.(((((((.......)))))))..)))))).)))............. (-20.11 = -20.30 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:46 2006