
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,874,558 – 9,874,694 |
| Length | 136 |
| Max. P | 0.993932 |

| Location | 9,874,558 – 9,874,670 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
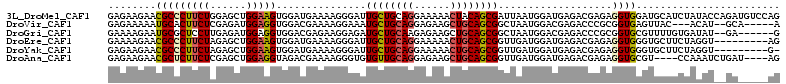
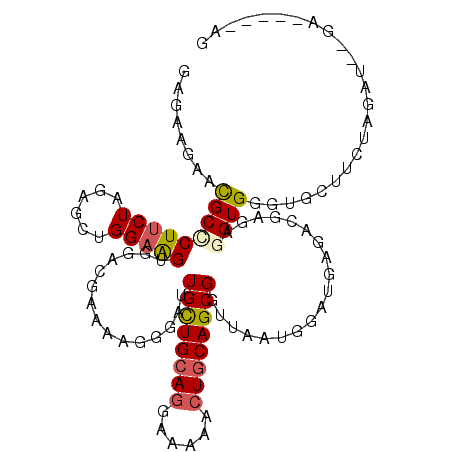
>3L_DroMel_CAF1 9874558 112 + 23771897 GAGAAGAACGCCCUUCUGGAGCUGGAAGUGGAUGAAAAGGGAUUGCUGCAGGAAAAACUACAGCGAUUAAUGGAUGAGACGAGAGGUGGAUGCAUCUAUACCAGAUGUCCAG .(((((......)))))(((.((((..(((((((......((((((((.((......)).))))))))...........(....).......))))))).))))...))).. ( -32.10) >DroVir_CAF1 32902 102 + 1 GAGAAAAAUGCACUUCUCGAGAUGGAGGUGGACGAAAAGGAAAUGCUGCAGGAGAAGCUGCAGCGGCUAAUGGACGAGACCCGCGGUGAGUUAC---ACAU--GCA-----A ..........(((((((......)))))))........((...((((((((......)))))))).))...((.......))((((((.....)---)).)--)).-----. ( -27.10) >DroGri_CAF1 34498 104 + 1 GAAAAGAAUGCGCUCCUUGAGAUGGAGGUGGACGAGAAGGAGAUGCUGCAAGAGAAGCUGCAGCGGCUAAUGGACGAGACCCGCGGUGCGUUUUGUGAUAU--GA------G ....((((((((((((.......))).((((.((...(..((.(((((((........))))))).))..)...))....)))).))))))))).......--..------. ( -28.50) >DroEre_CAF1 37686 103 + 1 GAAAAGAACGCCCUUCUAGAGCUGGAAGUGGAUGAAAAGGGAUUGCUGCAGGAAAAACUGCAGCGGUUGAUGGAUGAGACGAGAGGUGGGUGCUUCUAGGU---------AG ........((((((((((....)))))).)).))......(((((((((((......)))))))))))..........((.((((((....))))))..))---------.. ( -32.00) >DroYak_CAF1 36620 102 + 1 GAGAAGAACGCCCUUCUAGAGCUGGAAGUGGAUGAAAAGGGAUUGCUGCAGGAAAAACUGCAGCGGUUGAUGGAUGAGACGAGAGGUGGGUGCUUCUAGGU---------G- .(((((..((((((((((....))))))............(((((((((((......)))))))))))................))))....)))))....---------.- ( -33.30) >DroAna_CAF1 34602 104 + 1 GAGAAGAACGCUCUUCUCGAGCUGGAGGUAGACGAAAAGGGUGUGUUGCAGGAGAAGCUGCAGCGGUUGAUGGAUGAGACGAGAGGUGCGU----CCAAAUCUGAU----AG .......((((((((.(((..(((....))).))).))))))))(((((((......)))))))((((..((((((...(....)...)))----)))))))....----.. ( -33.90) >consensus GAGAAGAACGCCCUUCUAGAGCUGGAAGUGGACGAAAAGGGAUUGCUGCAGGAAAAACUGCAGCGGUUAAUGGAUGAGACGAGAGGUGGGUGCUUCUAGAU__GA_____AG ........(((((((((......)))))...............((((((((......))))))))...................))))........................ (-16.59 = -16.73 + 0.14)

| Location | 9,874,558 – 9,874,670 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -13.71 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9874558 112 - 23771897 CUGGACAUCUGGUAUAGAUGCAUCCACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCCAGAAGGGCGUUCUUCUC (((((....(((...(((((...........)))))...)))......(((((((......)))))))..................)))))....((((((....)))))). ( -24.80) >DroVir_CAF1 32902 102 - 1 U-----UGC--AUGU---GUAACUCACCGCGGGUCUCGUCCAUUAGCCGCUGCAGCUUCUCCUGCAGCAUUUCCUUUUCGUCCACCUCCAUCUCGAGAAGUGCAUUUUUCUC .-----(((--(.((---(.....))).((.((......))....)).(((((((......))))))).....(((((((.............)))))))))))........ ( -24.42) >DroGri_CAF1 34498 104 - 1 C------UC--AUAUCACAAAACGCACCGCGGGUCUCGUCCAUUAGCCGCUGCAGCUUCUCUUGCAGCAUCUCCUUCUCGUCCACCUCCAUCUCAAGGAGCGCAUUCUUUUC .------..--.................(((((...((......((..(((((((......)))))))..))......)).))..((((.......)))))))......... ( -21.30) >DroEre_CAF1 37686 103 - 1 CU---------ACCUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCUAGAAGGGCGUUCUUUUC ..---------....(((((.((((.......................(((((((......)))))))................((((........)))))).)).))))). ( -21.10) >DroYak_CAF1 36620 102 - 1 -C---------ACCUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUCCCUUUUCAUCCACUUCCAGCUCUAGAAGGGCGUUCUUCUC -.---------....(((((.((((.......................(((((((......)))))))................((((........)))))).)).))))). ( -23.70) >DroAna_CAF1 34602 104 - 1 CU----AUCAGAUUUGG----ACGCACCUCUCGUCUCAUCCAUCAACCGCUGCAGCUUCUCCUGCAACACACCCUUUUCGUCUACCUCCAGCUCGAGAAGAGCGUUCUUCUC ..----...(((...((----((((.......................(.(((((......))))).).....(((((((.((......))..))))))).)))))).))). ( -23.20) >consensus CU_____UC__AUAUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGCUUCUCCUGCAGCAAUCCCUUUUCAUCCACCUCCAGCUCGAGAAGGGCGUUCUUCUC ................................................(((((((......)))))))...........................(((((......))))). (-13.71 = -13.10 + -0.61)


| Location | 9,874,598 – 9,874,694 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.21 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -9.88 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9874598 96 + 23771897 GAUUGCUGCAGGAAAAACUACAGCGAUUAAUGGAUGAGACGAGAGGUGGAUGCAUCUAUACCAGAUGUCCAGCACAGACU--U--UAAAACUAAUUCAUC ((((((((.((......)).))))))))....((((((.(....).(((..((((((.....))))))))).........--.--.........)))))) ( -23.40) >DroSec_CAF1 35559 95 + 1 GUUUGCUGCAGGAAAAACUACAGCGAUUAAUGGAUGAGACGAGAGGUGGAUGAUUCAAGACCAGACGUUCAGUACAAACU-----CAAAACUAAUUUAUC ..((((((.((......)).))))))........((((.(....).((((((..((.......)))))))).......))-----))............. ( -16.10) >DroSim_CAF1 37462 95 + 1 GUUUGCUGCAGGAGAAACUACAGCGAUUAAUGGAUGAGACGAGAGGUGGAUGAUUCAAGACCAGACGUUCAGUACAAACU-----CAAAACUAAUUCAUC ..((((((.((......)).))))))...((((((.((..(((.(((.((....))...)))..((.....)).....))-----)....)).)))))). ( -16.90) >DroEre_CAF1 37726 89 + 1 GAUUGCUGCAGGAAAAACUGCAGCGGUUGAUGGAUGAGACGAGAGGUGGGUGCUUCUAGGU---------AGUACAGACUAAUUCA--AACUAAUUCAUC (((((((((((......)))))))))))(((((((.((.(.((((((....)))))).).(---------(((....)))).....--..)).))))))) ( -30.50) >DroYak_CAF1 36660 87 + 1 GAUUGCUGCAGGAAAAACUGCAGCGGUUGAUGGAUGAGACGAGAGGUGGGUGCUUCUAGGU---------G----AAACCAAAUCAAAAACUAAUUCAUU (((((((((((......)))))))))))(((((((.((...((((((....)))))).((.---------.----...))..........)).))))))) ( -29.40) >DroAna_CAF1 34642 82 + 1 GUGUGUUGCAGGAGAAGCUGCAGCGGUUGAUGGAUGAGACGAGAGGUGCGU----CCAAAUCUGAU----AGUCUAAACU-----AUUA----AGUC-UU ....(((((((......)))))))((((..((((((...(....)...)))----)))))))((((----(((....)))-----))))----....-.. ( -25.40) >consensus GAUUGCUGCAGGAAAAACUACAGCGAUUAAUGGAUGAGACGAGAGGUGGAUGCUUCUAGACCAGA_____AGUACAAACU_____AAAAACUAAUUCAUC ..((((((.((......)).))))))......((((((.(....).((((....))))....................................)))))) ( -9.88 = -10.13 + 0.25)



| Location | 9,874,598 – 9,874,694 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.21 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -10.92 |
| Energy contribution | -10.83 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9874598 96 - 23771897 GAUGAAUUAGUUUUA--A--AGUCUGUGCUGGACAUCUGGUAUAGAUGCAUCCACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUUUCCUGCAGCAAUC (((((....((....--.--.((((((((..(....)..))))))))......))...))))).............(((((((......))))))).... ( -27.90) >DroSec_CAF1 35559 95 - 1 GAUAAAUUAGUUUUG-----AGUUUGUACUGAACGUCUGGUCUUGAAUCAUCCACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUUUCCUGCAGCAAAC .....((((((..((-----((..........(((...(((...((....)).)))...)))))))...)))))).(((((((......))))))).... ( -17.40) >DroSim_CAF1 37462 95 - 1 GAUGAAUUAGUUUUG-----AGUUUGUACUGAACGUCUGGUCUUGAAUCAUCCACCUCUCGUCUCAUCCAUUAAUCGCUGUAGUUUCUCCUGCAGCAAAC (((((.(((((....-----.......)))))(((...(((...((....)).)))...))).)))))........(((((((......))))))).... ( -19.90) >DroEre_CAF1 37726 89 - 1 GAUGAAUUAGUU--UGAAUUAGUCUGUACU---------ACCUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUC ((((...((((.--.((.....))...)))---------)....((..(...........)..))...))))....(((((((......))))))).... ( -18.60) >DroYak_CAF1 36660 87 - 1 AAUGAAUUAGUUUUUGAUUUGGUUU----C---------ACCUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAUC ...(((((((...))))))).((((----(---------.....)))))...........................(((((((......))))))).... ( -17.20) >DroAna_CAF1 34642 82 - 1 AA-GACU----UAAU-----AGUUUAGACU----AUCAGAUUUGG----ACGCACCUCUCGUCUCAUCCAUCAACCGCUGCAGCUUCUCCUGCAACACAC .(-((((----....-----))))).....----....(((.(((----(((.......)))).))...)))....(.(((((......))))).).... ( -13.00) >consensus GAUGAAUUAGUUUUU_____AGUUUGUACU_____UCUGAUCUAGAAGCACCCACCUCUCGUCUCAUCCAUCAACCGCUGCAGUUUUUCCUGCAGCAAAC ............................................................................(((((((......))))))).... (-10.92 = -10.83 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:38 2006