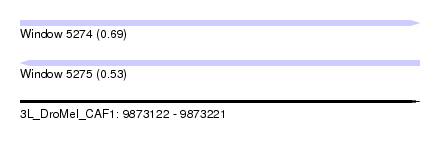
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,873,122 – 9,873,221 |
| Length | 99 |
| Max. P | 0.694852 |

| Location | 9,873,122 – 9,873,221 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -17.59 |
| Energy contribution | -19.76 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
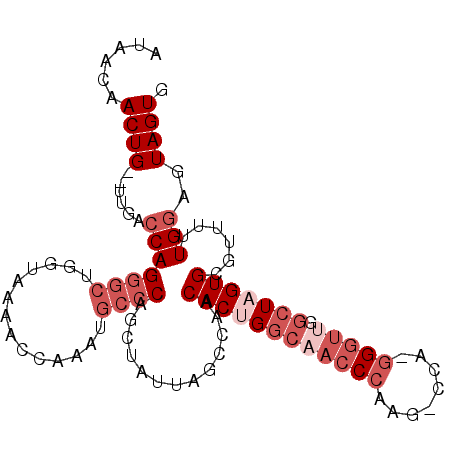
>3L_DroMel_CAF1 9873122 99 + 23771897 AUAACAACUG--UUGACCAGGGCUGGAAACGGCAAAUGCCCAUCUAUUGGCCAACACUGGCAACCCAAG-CCA-GGGUUGGCUUGUGCGUUUUUGGAGUAGUG .((((....)--))).((((.((((....))))((((((.((......(((((((.(((((.......)-)))-).))))))))).))))))))))....... ( -37.00) >DroVir_CAF1 31574 91 + 1 GUAUCGACUGUAUUGCACAGGGCUGGUUG--CCA------CGCGUACUUUACAACACUGCG---CCAGGGCUACAGGUUGGUUUGUGCAU-UUUGGAGUAGUG ((((((.((((.....))))(((.....)--)).------)).)))).......((((((.---((((.((.(((((....)))))))..-.)))).)))))) ( -27.00) >DroSec_CAF1 34094 99 + 1 AUAACAACUG--UUGACCAGGGCUGGUAAAACCAAAUGCCCAACAGUUAGCCAACACUGGCAACCCAAG-CCA-GGGUUGGCUAGUGCGUUUUUGGAGUAGUG ...((.(((.--(.((((((((((((.....)))...)))).....(((((((((.(((((.......)-)))-).))))))))))).)))...).))).)). ( -36.80) >DroSim_CAF1 35995 99 + 1 AUAUCAACUG--UUGACCAGGGCUGGUAAAACCAAAUGCCCAGCUAUUAGCCAACACUGGCAACCCAAG-CCA-GGGUUGGCUAGUGCGUUUUUGGAGUAGUG ......((((--....((((((((((.....)))...)))).((.((((((((((.(((((.......)-)))-).)))))))))))).....)))..)))). ( -40.30) >DroEre_CAF1 36132 99 + 1 AUAACAACUG--UUGGCCAGGGCUGUCAAAGCCAAAUGCCCAGUUAUUAUCCAACACUGGCUACCCAAG-CCA-GGGUUGGCUAGUGCGUUUUUGGAGUAGUG .(((.(((((--..(((...((((.....))))....)))))))).)))(((((((((((((((((...-...-))).))))))))).....)))))...... ( -37.00) >DroYak_CAF1 35082 99 + 1 AUAUCAACUG--UUGACCAGGGCUGUCAAAAUCAAAUGCCCGGCUAUUACUCAACACUGGCAACCCAAG-CCA-GGGUUGGCUAGUGCGUUUUUGGAGUAGUG ..........--....((.((((..............)))))).((((((((..((((((((((((...-...-))))).)))))))((....)))))))))) ( -31.94) >consensus AUAACAACUG__UUGACCAGGGCUGGUAAAACCAAAUGCCCAGCUAUUAGCCAACACUGGCAACCCAAG_CCA_GGGUUGGCUAGUGCGUUUUUGGAGUAGUG ......((((......(((((((..............)))).............((((((((((((........))))).)))))))......)))..)))). (-17.59 = -19.76 + 2.17)

| Location | 9,873,122 – 9,873,221 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -15.29 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
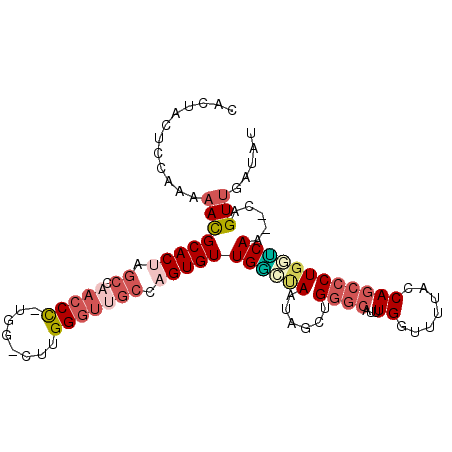
>3L_DroMel_CAF1 9873122 99 - 23771897 CACUACUCCAAAAACGCACAAGCCAACCC-UGG-CUUGGGUUGCCAGUGUUGGCCAAUAGAUGGGCAUUUGCCGUUUCCAGCCCUGGUCAA--CAGUUGUUAU ............(((((.(((((((....-)))-)))).((((((((.(((((.....((((((.(....)))))))))))).)))).)))--).).)))).. ( -32.90) >DroVir_CAF1 31574 91 - 1 CACUACUCCAAA-AUGCACAAACCAACCUGUAGCCCUGG---CGCAGUGUUGUAAAGUACGCG------UGG--CAACCAGCCCUGUGCAAUACAGUCGAUAC ............-.((((((...((((((((.((....)---))))).))))..........(------.((--(.....))))))))))............. ( -22.50) >DroSec_CAF1 34094 99 - 1 CACUACUCCAAAAACGCACUAGCCAACCC-UGG-CUUGGGUUGCCAGUGUUGGCUAACUGUUGGGCAUUUGGUUUUACCAGCCCUGGUCAA--CAGUUGUUAU ............(((.....(((((((.(-(((-(.......))))).)))))))(((((((((.((.((((.....))))...)).))))--)))))))).. ( -37.30) >DroSim_CAF1 35995 99 - 1 CACUACUCCAAAAACGCACUAGCCAACCC-UGG-CUUGGGUUGCCAGUGUUGGCUAAUAGCUGGGCAUUUGGUUUUACCAGCCCUGGUCAA--CAGUUGAUAU .(((...(((.....((..((((((((.(-(((-(.......))))).))))))))...)).((((...(((.....))))))))))....--.)))...... ( -35.00) >DroEre_CAF1 36132 99 - 1 CACUACUCCAAAAACGCACUAGCCAACCC-UGG-CUUGGGUAGCCAGUGUUGGAUAAUAACUGGGCAUUUGGCUUUGACAGCCCUGGCCAA--CAGUUGUUAU ......................(((((.(-(((-((.....)))))).)))))(((((((((((((....((((.....))))...)))..--)))))))))) ( -36.00) >DroYak_CAF1 35082 99 - 1 CACUACUCCAAAAACGCACUAGCCAACCC-UGG-CUUGGGUUGCCAGUGUUGAGUAAUAGCCGGGCAUUUGAUUUUGACAGCCCUGGUCAA--CAGUUGAUAU ...(((((.......(((((.((.(((((-...-...))))))).))))).)))))...(((((((..............)))).)))...--.......... ( -27.14) >consensus CACUACUCCAAAAACGCACUAGCCAACCC_UGG_CUUGGGUUGCCAGUGUUGGCUAAUAGCUGGGCAUUUGGUUUUACCAGCCCUGGUCAA__CAGUUGAUAU ............((((((((.((.(((((........))))))).)))))((((((......((((...((.......)))))))))))).....)))..... (-15.29 = -16.73 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:35 2006