
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,863,185 – 9,863,332 |
| Length | 147 |
| Max. P | 0.894714 |

| Location | 9,863,185 – 9,863,292 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.94 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
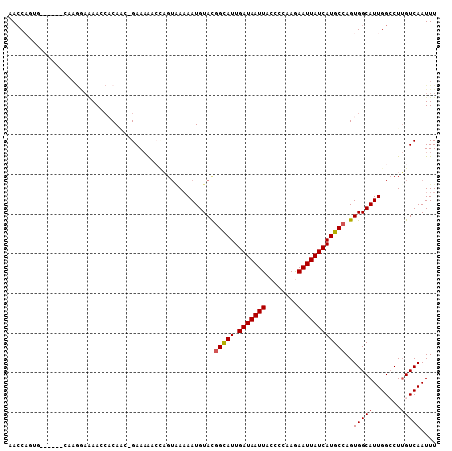
>3L_DroMel_CAF1 9863185 107 + 23771897 AACCAGUGCGUGGACGAGGAAAACCACAGC-GAAAAACCAGUAAAAAUGUACGGCAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUU ............(((((((..((((((...-.........(((......)))(((((.(((((((........)))))))))))).))))..))..)))))))..... ( -30.40) >DroSim_CAF1 24111 107 + 1 AACCAGUGCGUGGACGAGGAAAACCACAGU-GAAAAGCCAGUAAAAAUGUACGGCAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUU ............(((((((..((((((...-.........(((......)))(((((.(((((((........)))))))))))).))))..))..)))))))..... ( -30.40) >DroEre_CAF1 26153 107 + 1 AACCAGUGCGUGGAACAGGAAAAGCACAUC-GAAAAACCAGUGAAAAUGUACGGCAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUU ..((((((((.....)........(((...-.........(((......)))(((((.(((((((........)))))))))))).))))))))))............ ( -25.20) >DroWil_CAF1 45638 96 + 1 GAGC----------UAAAAAAAAACACAAUCGAACA--UUGUGAAAAUGUAUGGCAUUGAUAAUUACCCCAAAAAUUAUCAUGCGAGUGGCAUUGGGCUAGUCAAUUU .(((----------(.........((((((.....)--)))))..(((((.(.((((.(((((((........))))))))))).)...))))).))))......... ( -22.40) >DroAna_CAF1 23569 98 + 1 AACC----------CAAGAAGAAUCCCAACUGAAAGCCGAGUUAAAAUGUACGGCAUUGAUAAUUACCCCAAAAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUU ....----------.....................((((.((........))(((((.(((((((........))))))))))))..))))((((((...)))))).. ( -20.30) >DroPer_CAF1 22196 89 + 1 AACCAGAG-------------AAU-CCAUU-GAAAAU----AAAUAAUGUACGGUAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGACUUGUCAAUUU ........-------------...-..(((-((....----...(((((((((((((.(((((((........)))))))))))).)).))))))......))))).. ( -18.42) >consensus AACCAGUG______CAAGGAAAACCACAAC_GAAAAACCAGUAAAAAUGUACGGCAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUU ....................................................(((((.(((((((........))))))))))))..(((((.......))))).... (-14.74 = -14.93 + 0.19)


| Location | 9,863,219 – 9,863,332 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
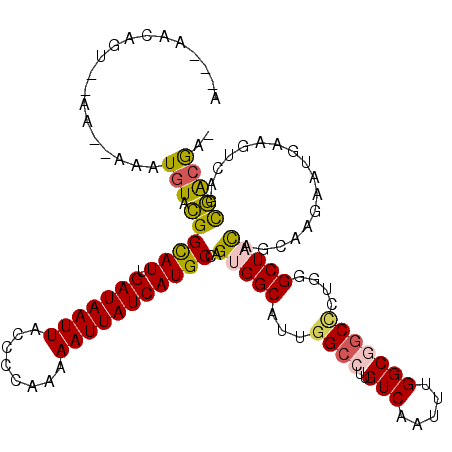
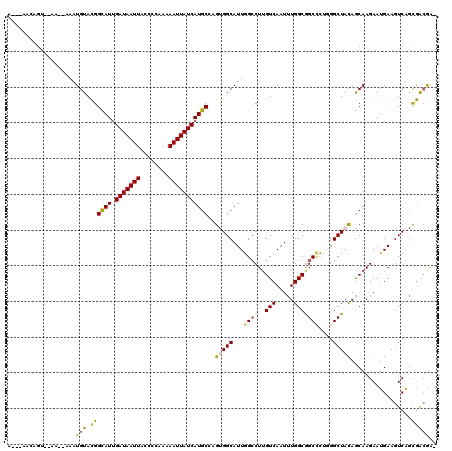
>3L_DroMel_CAF1 9863219 113 + 23771897 A---ACCAGU--AA--AAAUGUACGGCAUUGAUAAUUACCCCAAGAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUUGGCGGCCUUGGGCUACAGCAAGAAUGAAGUCAGCGGCGGG .---.((...--..--........(((((.(((((((........)))))))))))).(((((...((((..(((.....)))))))....))))).((.....((....))...)))). ( -34.20) >DroVir_CAF1 21408 116 + 1 A---AACAGUGAAAUAAAAUGUACGGCAUUGAUAAUUAUCCAAAAAAUUAUCAUGCGAGCGGCAUUGGCCUUGUCAAUCUGGCGGCCCUCGGCUACAGCAAGAAUGAAGUCAGCGACGA- .---...............(((.((((((.(((((((........)))))))))))(((.(((....(((..........))).))))))((((.((.......)).))))..))))).- ( -27.90) >DroGri_CAF1 22663 120 + 1 AGACAACCAAAAAAAAAUAUGUACGGCAUCGAUAAUUAUCCAAAAAAUUAUCAUGCGAGCGGCAUUGGCCUUGUCAAUCUGGCAGCGCUGGGCUACAGCAAGAAUGAAGUCAGCGACAAU .(((.....................((((.(((((((........)))))))))))..(((((....))).((((.....))))))((((.....)))).........)))......... ( -28.40) >DroWil_CAF1 45663 111 + 1 A-----UUGU--GA--AAAUGUAUGGCAUUGAUAAUUACCCCAAAAAUUAUCAUGCGAGUGGCAUUGGGCUAGUCAAUUUGGCGGCCUUGGGCUAUAGCAAGAAUGAAGUCAGCAGCAAU (-----((((--..--...(((.((((..((((((((........))))))))(((..(((((...(((((.(((.....))))))))...))))).)))........)))))))))))) ( -30.90) >DroMoj_CAF1 21849 114 + 1 G---AACUGUGCAAC--AAUGUACGGUAUUGAUAAUUAUCCCAAAAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUUGGCAGCUCUCGGCUACAACAAGAAUGAAGUCAGCGACGA- .---.((((((((..--..))))))))..((((((((........))))))))(((((.(((((.......)))))...)))))(((...((((.((.......)).))))))).....- ( -30.80) >DroAna_CAF1 23594 113 + 1 G---CCGAGU--UA--AAAUGUACGGCAUUGAUAAUUACCCCAAAAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUUGGCGGCCUUGGGCUACAGCAAGAACGAAAUCGAUGGAGGG .---((....--..--...(((..(((((.(((((((........)))))))))))).(((((...((((..(((.....)))))))....))))).)))....((....)).....)). ( -33.60) >consensus A___AACAGU__AA__AAAUGUACGGCAUUGAUAAUUACCCCAAAAAUUAUCAUGCCAGUGGCAUUGGCCUUGUCAAUUUGGCGGCCCUGGGCUACAGCAAGAAUGAAGUCAGCGACGA_ ...................(((.((((((.(((((((........)))))))))))..(((((...((((..(((.....)))))))....))))).................))))).. (-21.32 = -21.22 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:33 2006