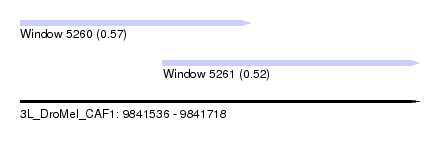
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,841,536 – 9,841,718 |
| Length | 182 |
| Max. P | 0.568283 |

| Location | 9,841,536 – 9,841,641 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -26.89 |
| Energy contribution | -26.86 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
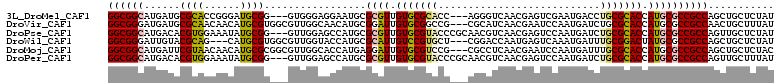
>3L_DroMel_CAF1 9841536 105 + 23771897 GGCGGCAUGAUGCGCACCGGGAUGCGG---GUGGGAGGAAUGCGCGUUGUGCGCACC---AGGGUCAACGAGUCGAAUGACCUGCGCACCAUGCGCCGCCAGCUGCUCUAU ((((((......(.((((.(....).)---))).).((...((((((.(((((((..---..(((((..........)))))))))))).))))))..)).)))))).... ( -47.30) >DroVir_CAF1 1504 108 + 1 GGCGGGAUGAUGCGCAACAACAUGCGUGGCGUUGGCAACAUGCGGAUUGUGCGGCCG---CGCAUCAACGAAUCCAAUGAUCUGCGCACCAUGCGCCGCCAACUGCUUUAU (((((((((.((((((.....(((((((((..((....))((((.....))))))))---)))))....((.((....)))))))))).))).).)))))........... ( -42.10) >DroPse_CAF1 1632 108 + 1 GGCGGCAUGACACGUGGAAAUAUGCGG---GUUGGAGCCAUGCGCGUUGUGCGUACCCGCAACGUCAACGAGUCCAAUGAUCUGCGCACCAUGCGCCGCCAGUUGCUCUAU ((((((..(((.(((.((....(((((---((.(....)((((((...)))))))))))))...)).))).)))...........((.....))))))))........... ( -40.20) >DroWil_CAF1 15728 105 + 1 GGCGGGAUUGUACGCAG---CAUGCGUGGCGUUGGUACCAUGCGCAUUGUCCGUGCU---CGGACCAAUGAGUCAAAUGAUUUGCGGACUAUGCGCCGCCAGCUGCUCUAU .(((........)))((---((.((.(((((.((....)).((((((.(((((((((---((......)))))..........)))))).))))))))))))))))).... ( -41.90) >DroMoj_CAF1 1485 108 + 1 GGCGGCAUGAUUCGUAACAACAUGCGCGGCGUUGGCACCAUGAGGAUUGUGCGUCCG---CGCCUCAACGAAUCCAAUGAUUUGCGCACCAUGCGCCGCCAGCUGCUCUAC ((((((..(((((((........(((((((((..(((((....))..)))))).)))---)))....)))))))...........((.....))))))))........... ( -39.40) >DroPer_CAF1 1635 108 + 1 GGCGGCAUGACACGUGGAAAUAUGCGG---GUUGGAGCCAUGCGCGUUGUGCGUACCCGCAACGUCAACGAGUCCAAUGAUCUGCGCACCAUGCGCCGCCAGUUGCUUUAU ((((((..(((.(((.((....(((((---((.(....)((((((...)))))))))))))...)).))).)))...........((.....))))))))........... ( -40.20) >consensus GGCGGCAUGAUACGCAGCAACAUGCGG___GUUGGAACCAUGCGCAUUGUGCGUACC___AGCGUCAACGAGUCCAAUGAUCUGCGCACCAUGCGCCGCCAGCUGCUCUAU ((((((......((((......)))).................((((.(((((((...........................))))))).))))))))))........... (-26.89 = -26.86 + -0.03)

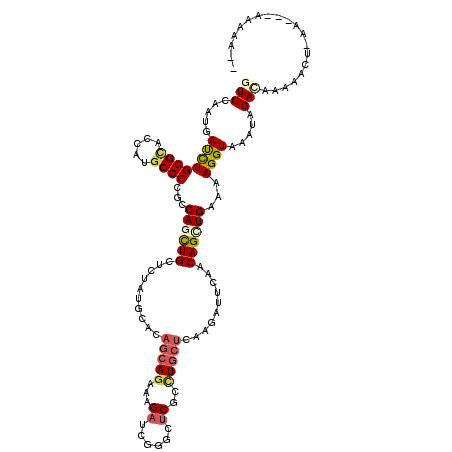
| Location | 9,841,601 – 9,841,718 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9841601 117 + 23771897 GUCGAAUGACCUGCGCACCAUGCGCCGCCAGCUGCUCUAUGCACAGCAGAAAGACCGGGCUCGCCUGCUCAAGAUUCAGCAGCUGAAAAGGUAAAUAGACCAAAUUUCAA---AAAAAAA (((.....((((((((.....))))...((((((((...(((...)))....((.((((....)))).)).......))))))))...)))).....)))..........---....... ( -35.30) >DroGri_CAF1 1656 115 + 1 AUCCAAUGAUCUGCGCACCAUGCGCCGCCAACUGCUAUAUGCACAGCAAAAAGAUCGCGCUCGCCUCCUCAAGAUUCAACAGCUGAAAAGGUAAUUAGACAAAAACA--AAAAAAAA--- .......(((((((((.....)))).((....(((.....)))..))....)))))......((((..(((............)))..))))...............--........--- ( -19.90) >DroWil_CAF1 15793 110 + 1 GUCAAAUGAUUUGCGGACUAUGCGCCGCCAGCUGCUCUAUGCUCAGCAGAAAGAUAAAGCUCUAUUGGCUAAAAUAGAGCAGCUGAAAAGGUAAUUAGAUACA-----AA---AACAA-- (((.(((.((((((((........))))((((((((((((....(((.((.(((......))).)).)))...))))))))))))...)))).))).)))...-----..---.....-- ( -33.20) >DroMoj_CAF1 1553 117 + 1 AUCCAAUGAUUUGCGCACCAUGCGCCGCCAGCUGCUCUACGCACAGCAGAAAGAUCGCGCUCGUCUGCUCAAGAUACAACAGUUGAAAAGGUAAUUAGACAAAAAGUUAAACAAACA--- ......((((((((((.....))))...((((((..........((((((.((......))..))))))..........))))))..................))))))........--- ( -21.75) >DroAna_CAF1 2259 117 + 1 GUCGAAUGACCUGCGUACCAUGCGCCGCCAACUGCUCUAUGCUCAGCAGAAAGAUCGGGCCCGCCUGCUUAAGAUUCAGCAGCUGAAAAGGUAAAUAGACAAAAACGAAA---UAAGAAA (((.....(((((((((....(((........)))..)))))(((((.........(((....)))(((........))).)))))..)))).....)))..........---....... ( -26.00) >DroPer_CAF1 1703 117 + 1 GUCCAAUGAUCUGCGCACCAUGCGCCGCCAGUUGCUUUAUGCACAGCAGAAAGAUCGGGCUCGCCUGCUUAAGAUUCAACAGCUGAAAAGGUAAAUAGACAAAAGACAAA---UAGAAAA (((...((.(((((((.....)))).(((...(((.....)))((((.....((((.(((......)))...)))).....))))....)))....)))))...)))...---....... ( -26.90) >consensus GUCCAAUGAUCUGCGCACCAUGCGCCGCCAGCUGCUCUAUGCACAGCAGAAAGAUCGGGCUCGCCUGCUCAAGAUUCAACAGCUGAAAAGGUAAAUAGACAAAAACU_AA___AAAAA__ (((.....((((((((.....))))...((((((..........(((((...((......))..)))))..........))))))...)))).....))).................... (-16.80 = -17.50 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:21 2006