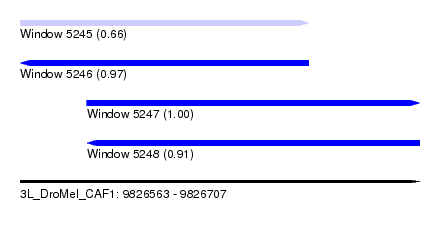
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,826,563 – 9,826,707 |
| Length | 144 |
| Max. P | 0.998423 |

| Location | 9,826,563 – 9,826,667 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9826563 104 + 23771897 ----------------UUGAUGGGCAAUAGAGGUGACACAGCAGCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUGGGAAAUGAAUAACUUUCAUUCUUCAAUCAUUAGAAA ----------------......((((....((.((......)).))....)))).(((((((...(((((((....)))))))((..((((((.....))))))..)).))))))).... ( -22.60) >DroSec_CAF1 120583 96 + 1 ---------------------GGGGAAUAAAGGUGACACA---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCGGAAA ---------------------..........(....)..(---((......)))....((((......)))).((((((((..(((((.((((.....))))))))).)))))))).... ( -19.00) >DroSim_CAF1 123821 116 + 1 AAGGGGAAGUUGGGGGUAGG-GGGGCAUUAAGGUGACACA---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCGGAAA ....((..((((((((..(.-.((((((((..(((.((((---.....)))).)))..)))))).(((((((....))))))).............))..)..))))))))..))..... ( -28.10) >DroEre_CAF1 123440 100 + 1 ----------------GUGG-GGGGUAUAAAGGUGACAUA---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCAGAAA ----------------....-...((((...(....)(((---((......)))))....)))).........((((((((..(((((.((((.....))))))))).)))))))).... ( -22.20) >DroYak_CAF1 133669 100 + 1 ----------------GUGG-GGGGUAUAAAGGUGACACA---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCGUCAGAAG ----------------....-...((((....(((.((((---.....)))).)))....)))).........((((((((..(((((.((((.....))))))))).)))))))).... ( -20.80) >consensus ________________GUGG_GGGGAAUAAAGGUGACACA___GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCAGAAA ................................(((.((((........)))).)))..((((......)))).((((((((..(((((.((((.....))))))))).)))))))).... (-18.00 = -17.12 + -0.88)

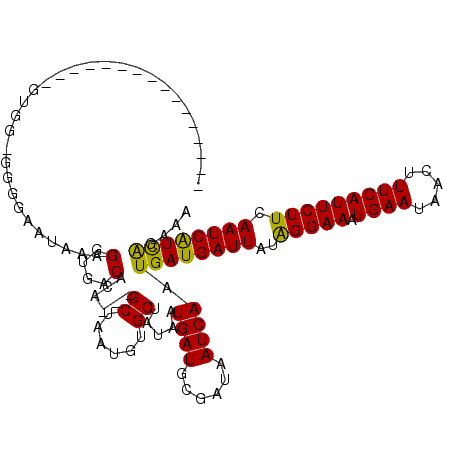

| Location | 9,826,563 – 9,826,667 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -18.17 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9826563 104 - 23771897 UUUCUAAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCCAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGCUGCUGUGUCACCUCUAUUGCCCAUCAA---------------- .....(((((((((.(((((((.....)))))))....((((((....))))))))).))))))((((((.......)))))).....................---------------- ( -17.00) >DroSec_CAF1 120583 96 - 1 UUUCCGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC---UGUGUCACCUUUAUUCCCC--------------------- .....((((((((.((((((((.....))))))..)).))))))))..((((......)))).((((((......))---))))...............--------------------- ( -18.10) >DroSim_CAF1 123821 116 - 1 UUUCCGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC---UGUGUCACCUUAAUGCCCC-CCUACCCCCAACUUCCCCUU .....((((((((.((((((((.....))))))..)).))))))))..........((((.......(((((.....---))))).......))))...-.................... ( -19.24) >DroEre_CAF1 123440 100 - 1 UUUCUGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC---UAUGUCACCUUUAUACCCC-CCAC---------------- ....(((((((((.((((((((.....))))))..)).))))))))).((((......)))).((((((......))---))))...............-....---------------- ( -18.40) >DroYak_CAF1 133669 100 - 1 CUUCUGACGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC---UGUGUCACCUUUAUACCCC-CCAC---------------- ....((((((((((.(((((((.....)))))))....((((((....))))))))).)))...(((((......))---)))))))............-....---------------- ( -18.10) >consensus UUUCUGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC___UGUGUCACCUUUAUACCCC_CCAA________________ .....(((((((((.(((((((.....)))))))....((((((....))))))))).))))))...(((((........)))))................................... (-14.18 = -14.42 + 0.24)

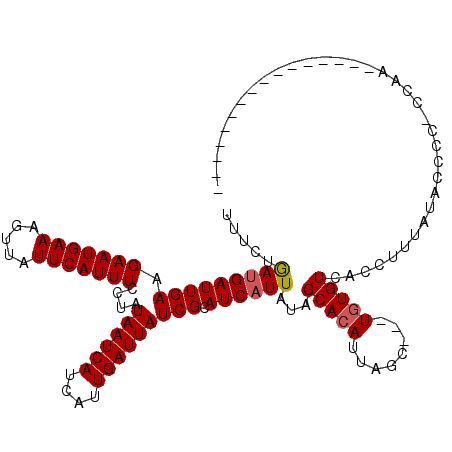

| Location | 9,826,587 – 9,826,707 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -24.98 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9826587 120 + 23771897 GCAGCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUGGGAAAUGAAUAACUUUCAUUCUUCAAUCAUUAGAAAACGUGAUCGUGAGCCGUUUGAAAUCGCAUUUCUAUAUCUU ..(((......)))(((..((((((((......(((((((..(((..((((((.....))))))..))))))))))..(((((.(..(....)))))))...))))))))..)))..... ( -27.90) >DroSec_CAF1 120602 117 + 1 ---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCGGAAAACGUGAUCGUGAGCCGUUACAAAUCGCAUUUCUAUAACUU ---((......))((((..((((((((.......(((((((..(((((.((((.....))))))))).)))))))((...(((....)))...)).......))))))))..)))).... ( -30.70) >DroSim_CAF1 123860 117 + 1 ---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCGGAAAACGUGAUCGUGAGCCGUUUCAAAUCGCAUUUCUAUAACUU ---((......))((((..((((((((.......(((((((..(((((.((((.....))))))))).)))))))((...(((....)))...)).......))))))))..)))).... ( -30.70) >DroEre_CAF1 123463 117 + 1 ---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCAGAAAACGUGUUUGUGUGCCAUUUAAAAUCGCAUUUCGAUAUGUU ---((......))(((...((((((((......((((((((..(((((.((((.....))))))))).))))))))......(..(....)..)........))))))))...))).... ( -24.90) >DroYak_CAF1 133692 117 + 1 ---GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCGUCAGAAGACGUGUAUGAGUGCCGUUGCAAAUCGCAUUUCGAUAUGUU ---((......))(((...((((((((......((((((((..(((((.((((.....))))))))).))))))))...((((.(((....)))))))....))))))))...))).... ( -29.10) >consensus ___GCUAAUGUGCUAUAAUGAUGCGAUAAUCAAUGAUGAUUAUAGGAAAUGAAUAACUUUCAUUCUUCAAUCAUCAGAAAACGUGAUCGUGAGCCGUUUCAAAUCGCAUUUCUAUAUCUU ...((......))((((..((((((((......((((((((..(((((.((((.....))))))))).))))))))...((((.(........)))))....))))))))..)))).... (-24.98 = -24.70 + -0.28)

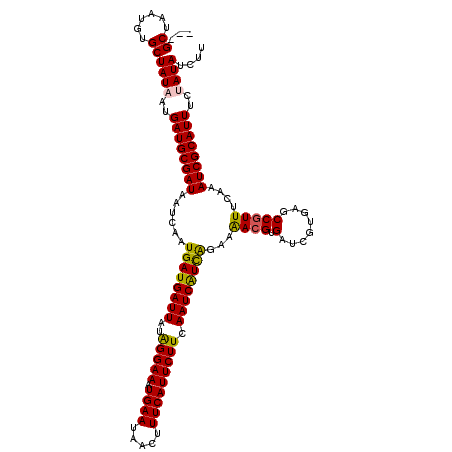
| Location | 9,826,587 – 9,826,707 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9826587 120 - 23771897 AAGAUAUAGAAAUGCGAUUUCAAACGGCUCACGAUCACGUUUUCUAAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCCAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGCUGC ....(((((..(((((((.((((.((.....)).............(((((((..(((((((.....)))))))....)))))))..))))..)))))))..)))))((......))... ( -23.00) >DroSec_CAF1 120602 117 - 1 AAGUUAUAGAAAUGCGAUUUGUAACGGCUCACGAUCACGUUUUCCGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC--- ..(((((((..(((((((...(((.((...(((....)))...))((((((((.((((((((.....))))))..)).)))))))).)))...)))))))..)))))))........--- ( -30.40) >DroSim_CAF1 123860 117 - 1 AAGUUAUAGAAAUGCGAUUUGAAACGGCUCACGAUCACGUUUUCCGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC--- ..(((((((..(((((((..((((((...........))))))..((((((((.((((((((.....))))))..)).)))))))).......)))))))..)))))))........--- ( -30.20) >DroEre_CAF1 123463 117 - 1 AACAUAUCGAAAUGCGAUUUUAAAUGGCACACAAACACGUUUUCUGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC--- ...........(((((((.((((.((.....))...........(((((((((.((((((((.....))))))..)).)))))))))))))..))))))).................--- ( -20.90) >DroYak_CAF1 133692 117 - 1 AACAUAUCGAAAUGCGAUUUGCAACGGCACUCAUACACGUCUUCUGACGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC--- ...........(((((((...(((.((...........(((....)))(((((.((((((((.....))))))..)).))))).)).)))...))))))).................--- ( -20.20) >consensus AAGAUAUAGAAAUGCGAUUUGAAACGGCUCACGAUCACGUUUUCUGAUGAUUGAAGAAUGAAAGUUAUUCAUUUCCUAUAAUCAUCAUUGAUUAUCGCAUCAUUAUAGCACAUUAGC___ ....(((((..(((((((...(((((...........)))))...((((((((..(((((((.....)))))))....)))))))).......)))))))..)))))............. (-19.56 = -20.80 + 1.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:56:08 2006