
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,824,162 – 9,824,361 |
| Length | 199 |
| Max. P | 0.999595 |

| Location | 9,824,162 – 9,824,282 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9824162 120 + 23771897 ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUACACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCA ............((((.((.(((((........(((....)))......)))))...)).))))(((....(((((((((.((..(((....)))..))..))))))))).....))).. ( -34.14) >DroSec_CAF1 118185 120 + 1 ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUAUACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCA ............((((.((.(((((........(((....)))......)))))...)).))))(((....(((((((((.((..(((....)))..))..))))))))).....))).. ( -32.24) >DroSim_CAF1 121427 120 + 1 ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUACAGUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCA ............((((((((((((..(((............)))....))))))))....))))(((....(((((((((.((..(((....)))..))..))))))))).....))).. ( -36.50) >DroEre_CAF1 120973 120 + 1 ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUUCACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCA ............((((.((...(((........(((....)))........)))...)).))))(((....(((((((((.((..(((....)))..))..))))))))).....))).. ( -33.19) >DroYak_CAF1 131189 120 + 1 ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUACACUUUGUAUGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAUCCAAAUUGGCCUUGGCCA ............(((....((.((((((((...(((((.......)))))........)))))))).))..((((((((..((..(((....)))..))...)))))))).....))).. ( -30.50) >consensus ACACAAAAACUCGCCUGACUGUGUGCCAUAAAAAUUUAAUAAUGAUAAAUACACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCA ............((((.((.(((((........(((....)))......)))))...)).))))(((....(((((((((.((..(((....)))..))..))))))))).....))).. (-30.14 = -30.78 + 0.64)

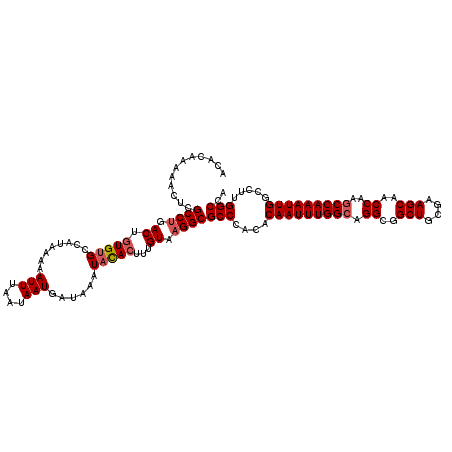

| Location | 9,824,162 – 9,824,282 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9824162 120 - 23771897 UGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGUAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ..(((...(((((((((((.(((((((...)))))))...))))))))).)).)))((((.((...(((((((((.......))))).......))))...)).))))............ ( -37.61) >DroSec_CAF1 118185 120 - 1 UGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUAUAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ..(((...(((((((((((.(((((((...)))))))...))))))))).)).)))(((.((.(((..(((........)))....))).)).)))((((((............)))))) ( -35.40) >DroSim_CAF1 121427 120 - 1 UGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAACUGUAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ..(((...(((((((((((.(((((((...)))))))...))))))))).)).)))((((.....(((((.....((((............))))...))))).))))............ ( -35.80) >DroEre_CAF1 120973 120 - 1 UGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGAAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ..(((...(((((((((((.(((((((...)))))))...))))))))).)).)))((((.((..(((((.....)))))............((.....)))).))))............ ( -37.50) >DroYak_CAF1 131189 120 - 1 UGGCCAAGGCCAAUUUGGAUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCAUACAAAGUGUAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ((((....))))((..(((((((((((...))))))(((((....(((((((....((((((.(((..(((........)))....))).)))))))))))))))))).)))))..)).. ( -37.10) >consensus UGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGUAUUUAUCAUUAUUAAAUUUUUAUGGCACACAGUCAGGCGAGUUUUUGUGU ..(((...(((((((((((.(((((((...)))))))...))))))))).)).)))(((...........(((((.......))))).....((((.....)))))))............ (-34.06 = -34.26 + 0.20)

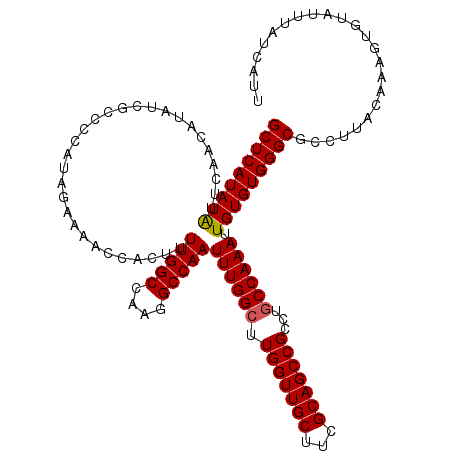
| Location | 9,824,202 – 9,824,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9824202 120 + 23771897 AAUGAUAAAUACACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCAAAAGUGGUUUUCUAUGGGGCGAUAUGUUGAAUAUAUGAGC .........(((.....)))...(((((.(((((((((((.((..(((....)))..))..))))))))(((....)))....)))..........)))))((((((...)))))).... ( -34.20) >DroSec_CAF1 118225 120 + 1 AAUGAUAAAUAUACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCAAAAGUGGUUUUCUAUGAGGCGAUAUGUUGAAUAUAUGAGC .......((((((.....(((((((....).(((((((((.((..(((....)))..))..)))))))))))))))(((....((((....))))..)))..))))))............ ( -33.30) >DroSim_CAF1 121467 120 + 1 AAUGAUAAAUACAGUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCAAAAGUGGUUUUCUAUGGGGCGAUAUGUUGAAUAUAUGAGC .............(((..((...(((((.(((((((((((.((..(((....)))..))..))))))))(((....)))....)))..........))))).(((.....)))))..))) ( -35.30) >DroEre_CAF1 121013 120 + 1 AAUGAUAAAUUCACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCAAAAGUGCUUUUCCAUGCGGCGAUAUGUUGAACAUAUGAGC .(((..(((..((((((.(((((((....).(((((((((.((..(((....)))..))..)))))))))))))))....))))))..))).)))...((.(((((.....)))))..)) ( -34.70) >DroYak_CAF1 131229 120 + 1 AAUGAUAAAUACACUUUGUAUGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAUCCAAAUUGGCCUUGGCCAAAAGUGGUUUUCUAUGCGGCGAUAUGUUGAACAUAUGAGC ................(((((((...((((..(((((((..((..(((....)))..))...)))))))(((....)))....))))....)))))))((.(((((.....)))))..)) ( -33.90) >consensus AAUGAUAAAUACACUUUGUAAGGCGCCCACACAAUUUGGCAGGCGGCUGCGAAGCAACCAAGCCAAAUUGGCCUUGGCCAAAAGUGGUUUUCUAUGCGGCGAUAUGUUGAAUAUAUGAGC ..................(((((((....).(((((((((.((..(((....)))..))..)))))))))))))))(((....((((....))))..))).((((((...)))))).... (-30.30 = -30.50 + 0.20)



| Location | 9,824,202 – 9,824,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -37.01 |
| Consensus MFE | -33.16 |
| Energy contribution | -33.12 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9824202 120 - 23771897 GCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGUAUUUAUCAUU ....(((((((........((((((((((..........(((((....)))))((((((.(((((((...)))))))...))))))))))).)))))........)))))))........ ( -39.19) >DroSec_CAF1 118225 120 - 1 GCUCAUAUAUUCAACAUAUCGCCUCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUAUAUUUAUCAUU ....(((((((........((((.((((.((........(((((....)))))((((((.(((((((...)))))))...)))))))).))))))))........)))))))........ ( -36.59) >DroSim_CAF1 121467 120 - 1 GCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAACUGUAUUUAUCAUU ....(((((..........((((((((((..........(((((....)))))((((((.(((((((...)))))))...))))))))))).)))))..........)))))........ ( -36.95) >DroEre_CAF1 121013 120 - 1 GCUCAUAUGUUCAACAUAUCGCCGCAUGGAAAAGCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGAAUUUAUCAUU ((..(((((.....))))).))...(((((((..((((((((((....))))....(((..((((((...))))))(((..((.....).)..))))))....))))))..))).)))). ( -38.20) >DroYak_CAF1 131229 120 - 1 GCUCAUAUGUUCAACAUAUCGCCGCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGAUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCAUACAAAGUGUAUUUAUCAUU ((..(((((.....))))).)).(.(((((....((((((.(((....)))(((((((..(((((((...)))))))....)))))))((((((...))))))))))))..))))))... ( -34.10) >consensus GCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCUGCCAAAUUGUGUGGGCGCCUUACAAAGUGUAUUUAUCAUU (((((((((..............................(((((....)))))((((((.(((((((...)))))))...)))))).)))))))))........................ (-33.16 = -33.12 + -0.04)


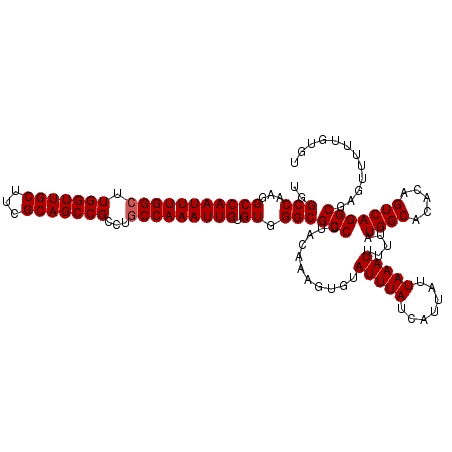
| Location | 9,824,242 – 9,824,361 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9824242 119 - 23771897 ACAAUAAAAA-AAAGCGUUGUCAACCGCUGAUGAUCCUUGGCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCU ..........-.(((.((..(((.....)))..)).)))(((...............................(((...(((((....)))))..)))...((((((...))))))))). ( -28.70) >DroSec_CAF1 118265 119 - 1 ACAAUAAAAA-AAAGCGUUGUCAACCGCUGAUGAUCCUUGGCUCAUAUAUUCAACAUAUCGCCUCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCU ..........-.(((.((..(((.....)))..)).)))(((...............................(((...(((((....)))))..)))...((((((...))))))))). ( -28.70) >DroSim_CAF1 121507 120 - 1 ACAAUAAAAAAAAAACGUUGUCAACCGAUGAUGAUCCUUGGCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCU ...............(((..((....))..)))......(((...............................(((...(((((....)))))..)))...((((((...))))))))). ( -28.20) >DroEre_CAF1 121053 111 - 1 ACC-----AA-AAAGCGUUGUCAGCCGCUG---AUCCUUGGCUCAUAUGUUCAACAUAUCGCCGCAUGGAAAAGCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCU ...-----..-...((......(((((((.---.(((.((((..(((((.....))))).))))...)))..)))....(((((....)))))...)))).((((((...)))))))).. ( -38.90) >DroYak_CAF1 131269 111 - 1 ACA-----AA-AAAGCGUUGUCAAGCGCUG---AUCCUUGGCUCAUAUGUUCAACAUAUCGCCGCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGAUUGGUUGCUUCGCAGCCGCCU ...-----..-...(((((((.((((((..---((((..(((..(((((.....))))).)))................(((((....)))))...))))..).))))).)))).))).. ( -37.30) >consensus ACAAUAAAAA_AAAGCGUUGUCAACCGCUGAUGAUCCUUGGCUCAUAUAUUCAACAUAUCGCCCCAUAGAAAACCACUUUUGGCCAAGGCCAAUUUGGCUUGGUUGCUUCGCAGCCGCCU .............((((........))))..........(((...............................(((...(((((....)))))..)))...((((((...))))))))). (-24.74 = -25.14 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:59 2006