
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,823,035 – 9,823,214 |
| Length | 179 |
| Max. P | 0.895727 |

| Location | 9,823,035 – 9,823,147 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9823035 112 + 23771897 GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCU----UUUU----UCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGUCUGUGUCUGUCUGGCUGUUUGU (((((..((((.....))))....)))))..(((----((((----(((.(((((....).)))))))))))((((..((((((((.((....)).))))))))..)))))))....... ( -32.40) >DroSec_CAF1 117065 104 + 1 GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCU----UUUU----UCCAGCUGCCUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU--------GUCUGGCUGUUUGU (((...((((((....(((.((((...((((.((----....----(((.((((.......))))))))).))))...)))))))...))))))....--------.))).......... ( -27.20) >DroSim_CAF1 120309 104 + 1 GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCU----UUUU----UCCAGCUGCCUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU--------GUCUGGCUGUUUGU (((...((((((....(((.((((...((((.((----....----(((.((((.......))))))))).))))...)))))))...))))))....--------.))).......... ( -27.20) >DroEre_CAF1 119819 109 + 1 GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCUGCUUUUUU----UCCAGCUGCU-UUGCCAGCGGAAGGGAGAUUUACACAGACAGAUCUUUGUG------UCUGUCUGGCUGUUUGU (((((..((((.....))))....)))))..((.((((((((----(((.((((..-....))))))))))))).......(((((((((......)------)))))))))).)).... ( -31.60) >DroYak_CAF1 130018 120 + 1 GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCUGCUUUUUUUUUCUCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACAGAUCUUUGUGUGUCUGUCUGUCUGGCUGUUUGU (((((..((((.....))))....)))))..((.(((.......(((((..(((((......)))))...))))).....((((((((((........))))))))))..))).)).... ( -34.50) >consensus GGAAAUAGAGAUAAAUUCUUGUGUUUUUCUUGCU____UUUU____UCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU_____UCUGUCUGGCUGUUUGU ......((((((....(((.((((...((((.((............(((.(((((....).))))))))).))))...)))))))...)))))).......................... (-23.86 = -23.62 + -0.24)



| Location | 9,823,035 – 9,823,147 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9823035 112 - 23771897 ACAAACAGCCAGACAGACACAGACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGA----AAAA----AGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC .......((.(((...((((((((...(....).))))))))...)))...((((((((.......)))).)))----)...----.))............................... ( -23.80) >DroSec_CAF1 117065 104 - 1 ACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAGGCAGCUGGA----AAAA----AGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC ..............--------....((((((...(((((((...(((..(((((((((.(...).)))).)))----....----))..)))...)))).)))....))))))...... ( -22.40) >DroSim_CAF1 120309 104 - 1 ACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAGGCAGCUGGA----AAAA----AGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC ..............--------....((((((...(((((((...(((..(((((((((.(...).)))).)))----....----))..)))...)))).)))....))))))...... ( -22.40) >DroEre_CAF1 119819 109 - 1 ACAAACAGCCAGACAGA------CACAAAGAUCUGUCUGUGUAAAUCUCCCUUCCGCUGGCAA-AGCAGCUGGA----AAAAAAGCAGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC .......((((((((((------........))))))))............((((((((....-..)))).)))----).....)).................................. ( -21.70) >DroYak_CAF1 130018 120 - 1 ACAAACAGCCAGACAGACAGACACACAAAGAUCUGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGAGAAAAAAAAAGCAGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC .......((.(.((((((((((.......).))))))))).)...(((((.....((((.......)))).)))))........)).................................. ( -24.10) >consensus ACAAACAGCCAGACAGA_____ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGA____AAAA____AGCAAGAAAAACACAAGAAUUUAUCUCUAUUUCC ..........................((((((...(((((((...(((..(((((((((.......)))).)))............))..)))...)))).)))....))))))...... (-17.80 = -18.20 + 0.40)

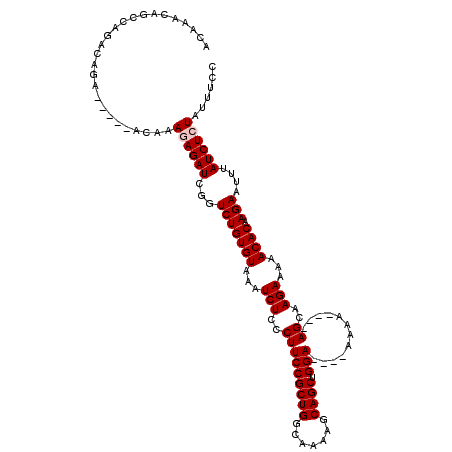
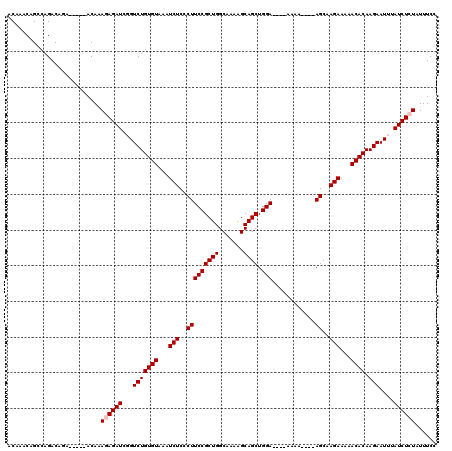
| Location | 9,823,071 – 9,823,185 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -36.74 |
| Consensus MFE | -27.31 |
| Energy contribution | -27.31 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9823071 114 + 23771897 UU----UCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGUCUGUGUCUGUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUG--UUUGC ..----...(((..((((((((.(((((((..((((..((((((((.((....)).))))))))..)))).(((((((....)))))))..))))).)))).))))).)...)--))... ( -37.00) >DroSec_CAF1 117101 108 + 1 UU----UCCAGCUGCCUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU--------GUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUGUGUUUGC ((----(((..((((........))))...)))))......((((((((((..((((.--------(((..(((((((....))))))).((.....)))))))))..))))).))))). ( -34.90) >DroSim_CAF1 120345 108 + 1 UU----UCCAGCUGCCUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU--------GUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUGUGUUUGC ((----(((..((((........))))...)))))......((((((((((..((((.--------(((..(((((((....))))))).((.....)))))))))..))))).))))). ( -34.90) >DroEre_CAF1 119859 107 + 1 UU----UCCAGCUGCU-UUGCCAGCGGAAGGGAGAUUUACACAGACAGAUCUUUGUG------UCUGUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUG--UUUGC ..----(((.((((..-....)))))))((((((.......(((((((((......)------))))))))(((((((....)))))))..))))))(((..(((.....)))--..))) ( -37.60) >DroYak_CAF1 130058 118 + 1 UUUUUCUCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACAGAUCUUUGUGUGUCUGUCUGUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUG--UUUGC ....(((((..(((((......)))))...))))).....((((((((((........)))))))))).(((((((((....)))).((((....))))))))).........--..... ( -39.30) >consensus UU____UCCAGCUGCUUUUGCCAGCGGAAGGGAGAUUUACACAGACCGAUCUCUUUGU_____UCUGUCUGGCUGUUUGUUUAAACGGCAGCUUCCUGCGGCCAAAAUGAUUG__UUUGC ..........(((((....).))))((.((((((.......(((((....................)))))(((((((....)))))))..))))))....))................. (-27.31 = -27.31 + 0.00)



| Location | 9,823,071 – 9,823,185 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -22.07 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9823071 114 - 23771897 GCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGACACAGACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGA----AA .....--.......((((((.((((....)))).((((....)))).))))))...((((((((...(....).)))))))).........((((((((.......)))).)))----). ( -34.60) >DroSec_CAF1 117101 108 - 1 GCAAACACAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAGGCAGCUGGA----AA ((.....(((.....)))...))....(((((((((((....)))).(((((..--------.....(((((..(......)..))))).......)))))...)))))))...----.. ( -29.04) >DroSim_CAF1 120345 108 - 1 GCAAACACAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAGGCAGCUGGA----AA ((.....(((.....)))...))....(((((((((((....)))).(((((..--------.....(((((..(......)..))))).......)))))...)))))))...----.. ( -29.04) >DroEre_CAF1 119859 107 - 1 GCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGA------CACAAAGAUCUGUCUGUGUAAAUCUCCCUUCCGCUGGCAA-AGCAGCUGGA----AA .....--........(((((.((((....)))).((((....)))).)))))(((((------((........)))))))...........((((((((....-..)))).)))----). ( -30.50) >DroYak_CAF1 130058 118 - 1 GCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGACAGACACACAAAGAUCUGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGAGAAAAA .....--........(((((.((((....)))).((((....)))).)))))((((((((((.......).))))))))).....(((((.....((((.......)))).))))).... ( -35.10) >consensus GCAAA__CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGA_____ACAAAGAGAUCGGUCUGUGUAAAUCUCCCUUCCGCUGGCAAAAGCAGCUGGA____AA ((.....(((.....)))(((((.(((((..((.((((....)))).))(((((....................)))))...........))))))).)))....))............. (-22.07 = -22.07 + 0.00)

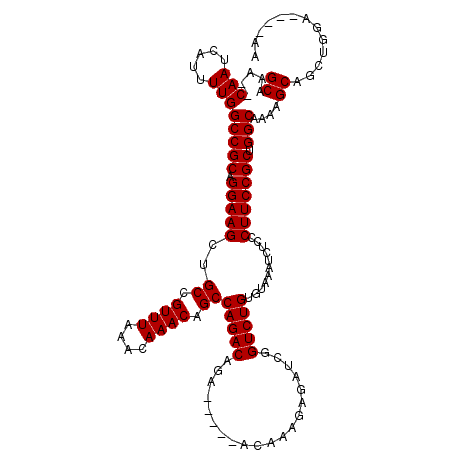
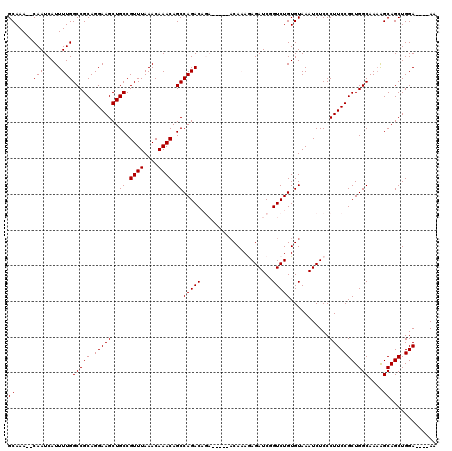
| Location | 9,823,107 – 9,823,214 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.98 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9823107 107 - 23771897 CACAAACUUAC-----------CAGCUUUCCGCCAAAAGUGCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGACACAGACAAAGAGAUCGGUCUGU ...((((....-----------((((((((.(((((((.((....--))....)))))))....))))))))..))))..............((((((...((........)).)))))) ( -25.50) >DroSec_CAF1 117137 104 - 1 CUCA-ACUUGCCAG-------CCAGCUUUCCGCCAAAAGUGCAAACACAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGU ....-....((..(-------.((((((((.((((((((((....))).....)))))))....)))))))).)((((....)))).))(((((--------............))))). ( -24.10) >DroSim_CAF1 120381 104 - 1 CUCA-ACUUACCAG-------CCAGCUUUCCGCCAAAAGUGCAAACACAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGAC--------ACAAAGAGAUCGGUCUGU ....-........(-------.((((((((.((((((((((....))).....)))))))....)))))))).)((((....))))...(((((--------............))))). ( -23.50) >DroEre_CAF1 119894 103 - 1 CUCA-CCUUGCCA--------CCACCUUUCCGCCAAAAGUGCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGA------CACAAAGAUCUGUCUGU ....-....((..--------..........(((((((.((....--))....))))))).((((....)))).((((....)))).))((((((((------........)))))))). ( -25.40) >DroYak_CAF1 130098 117 - 1 CUCA-CCUUACCACCUUAACACCACCUUUCCGCCAAAAGUGCAAA--CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGACAGACACACAAAGAUCUGUCUGU ....-..........................(((((((.((....--))....))))))).((((....)))).((((....))))......((((((((((.......).))))))))) ( -25.90) >consensus CUCA_ACUUACCA________CCAGCUUUCCGCCAAAAGUGCAAA__CAAUCAUUUUGGCCGCAGGAAGCUGCCGUUUAAACAAACAGCCAGACAGA_____ACAAAGAGAUCGGUCUGU ......................((((((((.(((((((.((......))....)))))))....))))))))..((((....))))...(((((....................))))). (-18.85 = -19.25 + 0.40)

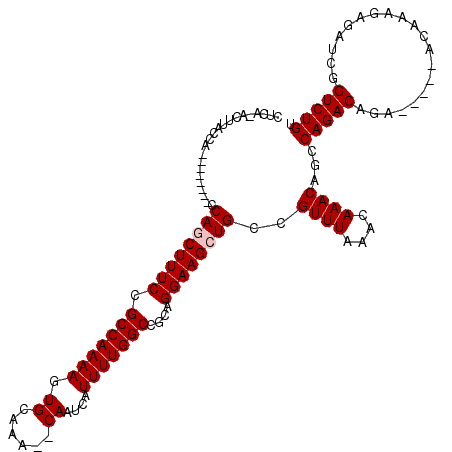
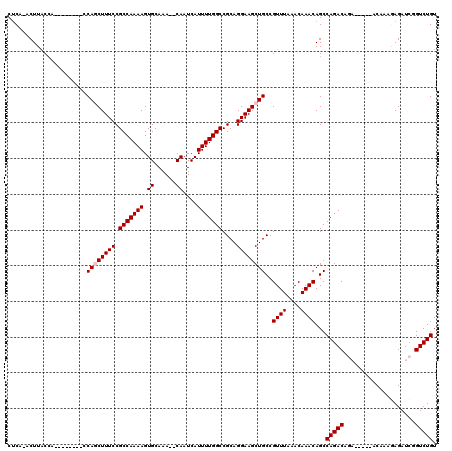
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:52 2006