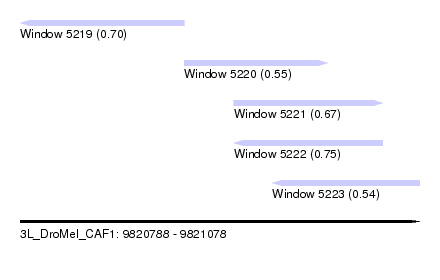
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,820,788 – 9,821,078 |
| Length | 290 |
| Max. P | 0.746557 |

| Location | 9,820,788 – 9,820,907 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.83 |
| Energy contribution | -29.95 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9820788 119 - 23771897 CAUACACACACACACACACACGCGGUACACCGCCCAGCUGUGUCAACAGGGGCGGAAAACG-GGCGGGGCCAUCCGGUCCUCAGCUUAGAAGGUCUUUUUUUUAAGCUGCAAUUCAUUCU ....................(((.((...((((((..((((....))))))))))...)).-.)))(((((....))))).((((((((((((....))))))))))))........... ( -39.50) >DroSim_CAF1 118068 110 - 1 CACACACGAACACACACACACGCGGUCCACCGCCCAGCUGUGUCAACAGGGGCGUAAAACGGGGCGGGGCCAUCCGGUCCUCAGCUUAGAAGGCUUUUUUUCAAAGCUAC---------- .......((...........(((((....))((((..((((....))))))))..........)))(((((....)))))))(((((.(((((....))))).)))))..---------- ( -32.50) >DroEre_CAF1 117757 111 - 1 UACA------CACACACACACGCAGUCCACCGCCCAGCUGUGUCAACAGGGGCGGAAAACGGGGCGGGGCCAUCCGGUCCUCAGUCGAAAAGGCAAUUUUU---AGCAACCACUCAUAUU ....------...........((.((((.((((((..((((....))))))))))......))))((((((....))))))..(((.....))).......---.))............. ( -34.00) >DroYak_CAF1 127767 109 - 1 CACA--------CACACACACGCAGUCCACCGCCCAGCUGUGUCAACAGGGGCGGAAAACGGGGCGGGGCCAUCCGGUCCUCAGCUGAAAAGGCAACUUUU---AGCUACAACUCAUAUU ....--------........(((.((...((((((..((((....))))))))))...))...)))(((((....)))))..((((((((.(....)))))---))))............ ( -39.20) >consensus CACA______CACACACACACGCAGUCCACCGCCCAGCUGUGUCAACAGGGGCGGAAAACGGGGCGGGGCCAUCCGGUCCUCAGCUGAAAAGGCAAUUUUU___AGCUACAACUCAUAUU ....................(((.((...((((((..((((....))))))))))...))...)))(((((....)))))..((((.(((((....)))))...))))............ (-29.83 = -29.95 + 0.12)

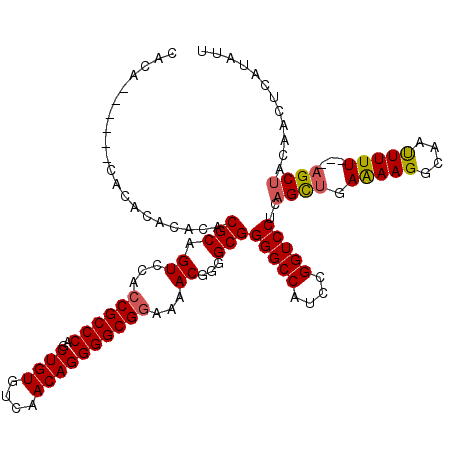

| Location | 9,820,907 – 9,821,011 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.71 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9820907 104 + 23771897 ----AGUGGGAGGUGAGGCUAUAUAGCUUCGAUUCGCCAUUUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA ----.....(((.(((((((....))))))).)))((((..((..(((------------(..(((((.((((((.((......))..)))))).))).))..))))..))...)))).. ( -34.20) >DroSec_CAF1 114844 100 + 1 AGUGAGUGGGAGGUGAGGCUAUAU--------UUCGCCAUUUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA .((.((...(((((((((((((((--------..(((.....))).))------------)))))).)))))))((((((((.(((.....))).))).))))).........)).)).. ( -31.20) >DroSim_CAF1 118178 96 + 1 ----AGUGGGAGGUGAGGCUAUAU--------UUCGCCAUUUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA ----.((.((((((((((((((((--------..(((.....))).))------------)))))).)))))))((((((((.(((.....))).))).)))))..........).)).. ( -30.80) >DroEre_CAF1 117868 111 + 1 ----AGUGAGAGGUGAGGCUUUAG--CUUCGAUUUGCAAUAUGUGUAUGUAUAUAUGUACGUAUCAAUCACCUCGCGACU---AUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA ----.(..((...((((((....)--))))).))..)....(((..(..(((((((((((((((.((((.((........---......))))))))))).))))))))))..)..))). ( -31.14) >DroYak_CAF1 127876 99 + 1 ----AGUGGGAGGUGAGGCUAUAG--CUACGAUACCACACAAGUGUGU------------GUAGCAAUCACCUCGCGACU---AUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA ----(((.(((((((((((....)--))..(.(((((((....)))).------------))).)..))))))).).)))---.((((((.(..((((((...))))))).))))))... ( -28.60) >consensus ____AGUGGGAGGUGAGGCUAUAU__CU_CGAUUCGCCAUUUGUGUAU____________GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAA ....((((.(((((((((((((............(((.....)))...............)))))).))))))).).)))....((((((.(..((((((...))))))).))))))... (-19.91 = -20.71 + 0.80)



| Location | 9,820,943 – 9,821,051 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9820943 108 + 23771897 UUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCACAGUUUCAC (((((...------------(((((...(((..(((((((((.(((.....))).))).))))))...))).((((......)))))))))....))))).................... ( -29.60) >DroSec_CAF1 114876 108 + 1 UUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCACAGUUUCAC (((((...------------(((((...(((..(((((((((.(((.....))).))).))))))...))).((((......)))))))))....))))).................... ( -29.60) >DroSim_CAF1 118206 108 + 1 UUGUGUAU------------GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCACAGUUUCAC (((((...------------(((((...(((..(((((((((.(((.....))).))).))))))...))).((((......)))))))))....))))).................... ( -29.60) >DroEre_CAF1 117902 117 + 1 AUGUGUAUGUAUAUAUGUACGUAUCAAUCACCUCGCGACU---AUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCACAGACGCAC .(((((.(((((((((((((((((.((((.((........---......))))))))))).)))))))))((((((......))))))......................))).))))). ( -30.04) >DroYak_CAF1 127910 105 + 1 AAGUGUGU------------GUAGCAAUCACCUCGCGACU---AUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCAUAGUCGCAC ..(.(.((------------(.......)))).)((((((---((.((((........))))(((..(((((((((((........))))......)))..))))..))))))))))).. ( -26.90) >consensus UUGUGUAU____________GUGGCUAUCACCUCGCGACUAUUAUCAAUGGGGUUAUACGUUGCGUAUGUGUGUUGGCAAAACAACGCCACUUUUCACAAUCACAAUCGCACAGUUUCAC .(((((..............((((((((.((((((.((......))..)))))).))).)))))((...((((.((((........)))).....))))...))....)))))....... (-22.18 = -22.58 + 0.40)


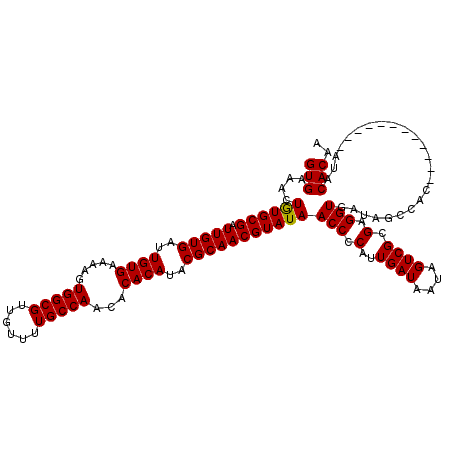
| Location | 9,820,943 – 9,821,051 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9820943 108 - 23771897 GUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAUAGUCGCGAGGUGAUAGCCAC------------AUACACAA .......((((...((((((((((....(((((.(((((.((((............))))...))))).)))))....))))))))))(((....)))..------------...)))). ( -28.10) >DroSec_CAF1 114876 108 - 1 GUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAUAGUCGCGAGGUGAUAGCCAC------------AUACACAA .......((((...((((((((((....(((((.(((((.((((............))))...))))).)))))....))))))))))(((....)))..------------...)))). ( -28.10) >DroSim_CAF1 118206 108 - 1 GUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAUAGUCGCGAGGUGAUAGCCAC------------AUACACAA .......((((...((((((((((....(((((.(((((.((((............))))...))))).)))))....))))))))))(((....)))..------------...)))). ( -28.10) >DroEre_CAF1 117902 117 - 1 GUGCGUCUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAU---AGUCGCGAGGUGAUUGAUACGUACAUAUAUACAUACACAU ((((((.((((((.(((((..((((.....(((((......)))))...))))..))))))))))).....((..((---..((....))..))..))))))))................ ( -33.70) >DroYak_CAF1 127910 105 - 1 GUGCGACUAUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAU---AGUCGCGAGGUGAUUGCUAC------------ACACACUU ..(((((((((((.(((((..((((.....(((((......)))))...))))..))))))))............))---))))))((((((........------------.))).))) ( -29.80) >consensus GUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAUAGUCGCGAGGUGAUAGCCAC____________AUACACAA (((....((((((.(((((..((((.....(((((......)))))...))))..)))))))))))(((.(..((((....)))).).)))........................))).. (-26.78 = -26.62 + -0.16)


| Location | 9,820,971 – 9,821,078 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9820971 107 - 23771897 CAA------------AAAGGCAAAAA-GCGAAGCGUGAGUGUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAU ...------------...((......-(((..(((((.(((((..(((.((((((....))))))...)))((((......))))..))))).)))))..)))......))......... ( -29.00) >DroSec_CAF1 114904 99 - 1 CAA--------------------AAA-GCGAAGCGUGAGUGUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAU ...--------------------...-(((..(((((.(((((..(((.((((((....))))))...)))((((......))))..))))).)))))..)))................. ( -27.70) >DroSim_CAF1 118234 99 - 1 CAA--------------------AAA-GCGAAGCGUGAGUGUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAU ...--------------------...-(((..(((((.(((((..(((.((((((....))))))...)))((((......))))..))))).)))))..)))................. ( -27.70) >DroEre_CAF1 117942 116 - 1 CAGUUUGCUUCUUUCUACGACAAAAA-GCGAAGCGAGAGUGUGCGUCUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAU--- ...((((((((...............-..))))))))((((.(.(..((((((.(((((..((((.....(((((......)))))...))))..)))))))))))..))))))...--- ( -30.93) >DroYak_CAF1 127938 117 - 1 CGGUUUGCUUGUUUCUACGACAAAAAGGCGAAGCGAGAGUGUGCGACUAUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAU--- ((.((((((((((.....))))....)))))).))..((((.(.(..((((((.(((((..((((.....(((((......)))))...))))..)))))))))))..))))))...--- ( -32.10) >consensus CAA_____________A_G_CAAAAA_GCGAAGCGUGAGUGUGAAACUGUGCGAUUGUGAUUGUGAAAAGUGGCGUUGUUUUGCCAACACACAUACGCAACGUAUAACCCCAUUGAUAAU ...........................(((..(((((.(((((...((.((((((....))))))...))(((((......))))).))))).)))))..)))................. (-24.30 = -24.70 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:44 2006