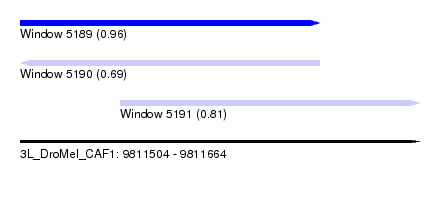
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,811,504 – 9,811,664 |
| Length | 160 |
| Max. P | 0.961249 |

| Location | 9,811,504 – 9,811,624 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -40.64 |
| Energy contribution | -40.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9811504 120 + 23771897 GCCUGUUCGGAGGUGAUGCGCUUGUUGGGAUCCAUUAGCAGCAGCUUCUGCAGCAGGUGAAAGGCCUUGCUGUCUGGCUUUAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUG ((((((((((((((..((((((...(((...)))..))).)))))))).(((((((((.....))).))))))((((((..((..((((......)))).))..))))))))))))))). ( -46.70) >DroSec_CAF1 105730 120 + 1 GCCUGUUCAGAGGUGAUGCGCUUGUUGGGAUCCAUUAGCAGCAGUUUCUGCAGCAGGUGAAAGGCCUUGCUGUCCGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUG ((((((((.(((((....((((((((((...))....((((......))))))))))))....)))))((((...(((..........)))...(((......))).)))))))))))). ( -43.40) >DroSim_CAF1 109040 120 + 1 GCCUGUUCAGAGGUGAUGCGCUUGUUGGGAUCCAUUAGCAGCAGUUUCUGUAGCAGAUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUACUUGGCCAGCGAGCAGGUG ((((((((.((((((((.(........).)))((((.((.((((...)))).)).))))....)))))...(.((((((.......((((........))))..))))))))))))))). ( -39.20) >DroEre_CAF1 108073 120 + 1 GCCUGUUCGGAGGUGAUGCGCUUGUUGGGGUCCAUUAGCAGUAGUUUCUGCAGCAGAUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUG (((((((((..(((((((((..((..(((((......((((......)))).(((((....(((((.........)))))...))))))))))..))))))))..)))..))))))))). ( -43.60) >DroYak_CAF1 118629 120 + 1 GCCUGUUCGGAGGUGAUGCGCUUGUUGGGGUCCAUUAGCAGCAGUUUCUGCAGCAGGUGGAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUG (((((((((..(((((((((..((..(((((((((..((.((((...)))).))..)))))(((((.........))))).........))))..))))))))..)))..))))))))). ( -46.00) >consensus GCCUGUUCGGAGGUGAUGCGCUUGUUGGGAUCCAUUAGCAGCAGUUUCUGCAGCAGGUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUG ((((((((.(((((....((((((((((...))....((((......))))))))))))....)))))((((...(((..........)))...(((......))).)))))))))))). (-40.64 = -40.88 + 0.24)

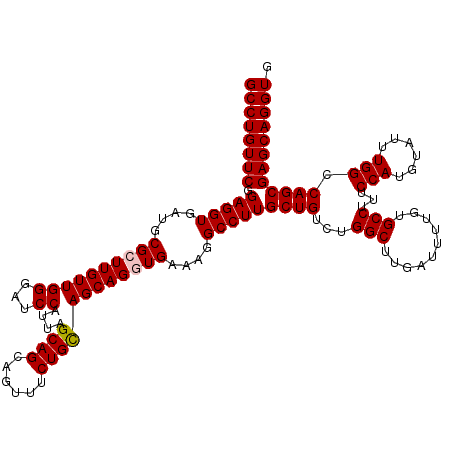

| Location | 9,811,504 – 9,811,624 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -27.82 |
| Energy contribution | -28.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9811504 120 - 23771897 CACCUGCUCGCUGGCCAAAUACAUGGAAAGGCACAAAAUAAAGCCAGACAGCAAGGCCUUUCACCUGCUGCAGAAGCUGCUGCUAAUGGAUCCCAACAAGCGCAUCACCUCCGAACAGGC ..((((.(((..(((((......)))...(((..........))).(.(((((.((.......)))))))).((.((.(((.....(((...)))...))))).)).))..))).)))). ( -30.60) >DroSec_CAF1 105730 120 - 1 CACCUGCUCGCUGGCCAAAUACAUGGAAAGGCACAAAAUCAAGCCGGACAGCAAGGCCUUUCACCUGCUGCAGAAACUGCUGCUAAUGGAUCCCAACAAGCGCAUCACCUCUGAACAGGC ..((((.((((((.(((......)))...(((..........)))...))))..((...((((...((.((((...)))).))...))))..))..................)).)))). ( -29.50) >DroSim_CAF1 109040 120 - 1 CACCUGCUCGCUGGCCAAGUACAUGGAAAGGCACAAAAUCAAGCCAGACAGCAAGGCCUUUCAUCUGCUACAGAAACUGCUGCUAAUGGAUCCCAACAAGCGCAUCACCUCUGAACAGGC ..((((.((((((.(((......)))...(((..........)))...))))..((...(((((..((..(((...)))..))..)))))..))..................)).)))). ( -28.60) >DroEre_CAF1 108073 120 - 1 CACCUGCUCGCUGGCCAAAUACAUGGAAAGGCACAAAAUCAAGCCAGACAGCAAGGCCUUUCAUCUGCUGCAGAAACUACUGCUAAUGGACCCCAACAAGCGCAUCACCUCCGAACAGGC ..((((.(((..((...........(((((((......((......)).......)))))))....(((((((......))))...(((...)))...)))......))..))).)))). ( -29.62) >DroYak_CAF1 118629 120 - 1 CACCUGCUCGCUGGCCAAAUACAUGGAAAGGCACAAAAUCAAGCCAGACAGCAAGGCCUUCCACCUGCUGCAGAAACUGCUGCUAAUGGACCCCAACAAGCGCAUCACCUCCGAACAGGC ..((((.((((((.(((......)))...(((..........)))...))))..((...((((...((.((((...)))).))...))))..))..................)).)))). ( -32.00) >consensus CACCUGCUCGCUGGCCAAAUACAUGGAAAGGCACAAAAUCAAGCCAGACAGCAAGGCCUUUCACCUGCUGCAGAAACUGCUGCUAAUGGAUCCCAACAAGCGCAUCACCUCCGAACAGGC ..((((.((((((.(((......)))...(((..........)))...))))..((...((((...((.((((...)))).))...))))..))..................)).)))). (-27.82 = -28.06 + 0.24)



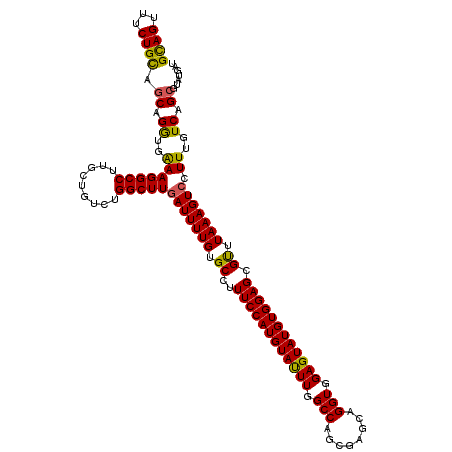
| Location | 9,811,544 – 9,811,664 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -36.32 |
| Energy contribution | -35.68 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9811544 120 + 23771897 GCAGCUUCUGCAGCAGGUGAAAGGCCUUGCUGUCUGGCUUUAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGUUUAAAGUCCUUUGUCAGCGUAUGAU ..((((...(((((((((.....))).))))))..)))).....(..(((...(((((((((((.(((........))).)))))))))))((...((((....))))...)))))..). ( -38.60) >DroSec_CAF1 105770 120 + 1 GCAGUUUCUGCAGCAGGUGAAAGGCCUUGCUGUCCGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGUUUAAAGUCCUUUGUCAGCGUAUGAU ((.......(((((((((.....))).))))))..(((..(((((((.((..((((((((((((.(((........))).)))))))))))).)).)))))))....))).))....... ( -39.50) >DroSim_CAF1 109080 120 + 1 GCAGUUUCUGUAGCAGAUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUACUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGUUUAAAGUCCUUUGUCAGCGUAUGAU ((((...)))).((.((..(((((((.........)))))(((((((.((..((((((((((((.(((........))).)))))))))))).)).))))))).))..)).))....... ( -38.20) >DroEre_CAF1 108113 120 + 1 GUAGUUUCUGCAGCAGAUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGUUUAAAGUCUUUUGUCAGCGUAUGGU ((((...))))............(((.(((.(.(((((..(((((((.((..((((((((((((.(((........))).)))))))))))).)).)))))))....))))))))).))) ( -37.10) >DroYak_CAF1 118669 120 + 1 GCAGUUUCUGCAGCAGGUGGAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGCUUAAAGUCCUUUGUCAGGGUAUGAU ((((...))))(((((((.....))).)))).((((((..(((((((.((..((((((((((((.(((........))).)))))))))))).)).)))))))....))))))....... ( -42.50) >consensus GCAGUUUCUGCAGCAGGUGAAAGGCCUUGCUGUCUGGCUUGAUUUUGUGCCUUUCCAUGUAUUUGGCCAGCGAGCAGGUGGAGUAUGUGGAGCGUUUAAAGUCCUUUGUCAGCGUAUGAU ((((...)))).((.((..(((((((.........)))))(((((((.((..((((((((((((.(((........))).)))))))))))).)).))))))).))..)).))....... (-36.32 = -35.68 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:13 2006