
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,800,918 – 9,801,049 |
| Length | 131 |
| Max. P | 0.989759 |

| Location | 9,800,918 – 9,801,033 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -9.44 |
| Energy contribution | -8.96 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
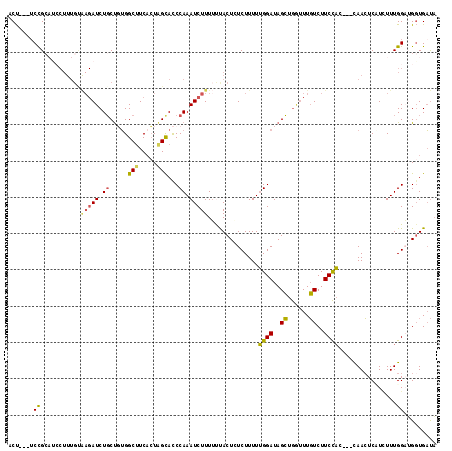
>3L_DroMel_CAF1 9800918 115 - 23771897 ACCGCAUCCGCAUCCUUUGUAAGAUCUGCUGGGGCUUCACUAGUACCCAAAUCUUUUUUACUCUCUUUUUGGAUAGCUGAUUUGUCUUCCAC---CAACUCCUCUUUGGAUGCUGCUA ...(((...((((((...((((((.....(((((((.....))).))))......))))))........((((..((......))..)))).---............))))))))).. ( -28.00) >DroSec_CAF1 95444 112 - 1 UCU---UCCGAACACUUUGUAAGAUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUAUCUUUUUGGAUAGCUGGUUUGUCUUCCAG---CAACUCAUCUUUGGAAGGUGAUU (((---(((((((.....))..((..(((((.(((...((((((..(((((................)))))...))))))..)))...)))---))..))....))))))))..... ( -28.99) >DroSim_CAF1 98223 100 - 1 ACU---GCCGC------------AUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUAUCUUUUUGGAUAGCUGGUUUGUCUUCCAG---CAACUCAUCUUUGGAAGGUGAUA ...---(((((------------(.....))))))...((((((..(((((................)))))...))))))(..((((((((---..........))))))))..).. ( -27.69) >DroEre_CAF1 97309 112 - 1 ACU---UCCGCAUCCUUUGCAAGAUCUGCCUUGGCCUCCAUGGCAUCCAAAUCUUUUCCACUCUCUUUUUGGAUAGCUGGUUUGUCUUCCAC---CAACUCCUUUUUGGCUGGUGAUA ...---...(((.....)))(((((..(((...(((.....)))(((((((................)))))))....)))..))))).(((---((...((.....)).)))))... ( -27.99) >DroYak_CAF1 107498 115 - 1 CCU---UCUGCUUCCUUUGUGAGAUCUUCUUUGGCUUCACUUGCACCCAAAUCUUUGCCACUCUCUUUUAGGAUAGAUACCUUUUCUUCUUCCCGUAAAUCUUGUUUGGCUGGUGACA ...---............(((((..(......)..))))).((((((.........((((..(...((((((..(((..........)))..)).))))....)..)))).)))).)) ( -18.90) >consensus ACU___UCCGCAUCCUUUGUAAGAUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUCUCUUUUUGGAUAGCUGGUUUGUCUUCCAC___CAACUCAUCUUUGGAUGGUGAUA .......((...........(((((.((.....(((.....)))...)).)))))..............((((..((......))..))))................))......... ( -9.44 = -8.96 + -0.48)


| Location | 9,800,955 – 9,801,049 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -16.27 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.49 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
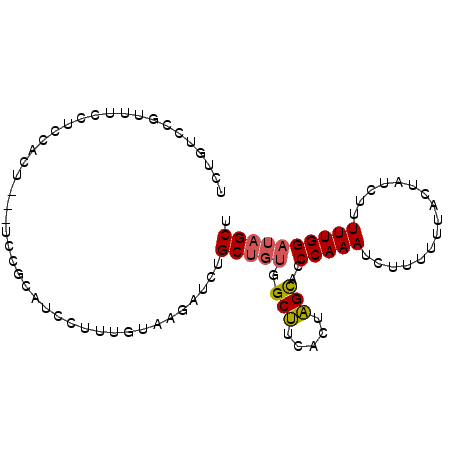
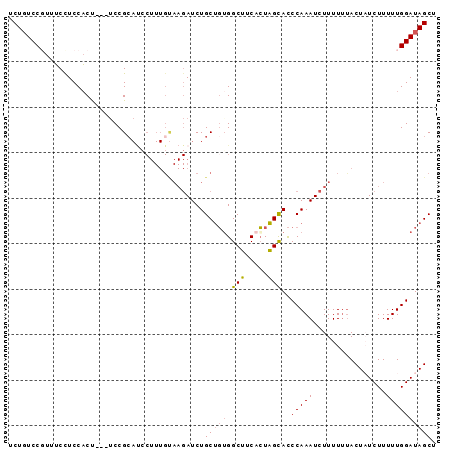
>3L_DroMel_CAF1 9800955 94 - 23771897 UCUCUCCGUUUCCUCCACCGCAUCCGCAUCCUUUGUAAGAUCUGCUGGGGCUUCACUAGUACCCAAAUCUUUUUUACUCUCUUUUUGGAUAGCU ...................(((((((........((((((.....(((((((.....))).))))......))))))........))))).)). ( -17.09) >DroSec_CAF1 95481 91 - 1 UCUGUCCGUUGCUUCCUCU---UCCGAACACUUUGUAAGAUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUAUCUUUUUGGAUAGCU .(((((((........(((---(.(((.....))).))))..(((((((....))).))))........................))))))).. ( -15.60) >DroSim_CAF1 98260 79 - 1 UCUGUCCCUUUCCUCCACU---GCCGC------------AUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUAUCUUUUUGGAUAGCU ...................---(((((------------(.....)))))).........................((((((....)))))).. ( -13.70) >DroEre_CAF1 97346 91 - 1 UCUGUCCUUUUCCUCCACU---UCCGCAUCCUUUGCAAGAUCUGCCUUGGCCUCCAUGGCAUCCAAAUCUUUUCCACUCUCUUUUUGGAUAGCU .((((((............---...(((.....)))(((((.((((.(((...))).)))).....)))))...............)))))).. ( -18.70) >consensus UCUGUCCGUUUCCUCCACU___UCCGCAUCCUUUGUAAGAUCUGCUGUGGCUUCACUAGCACCCAAAUCUUUUUUACUAUCUUUUUGGAUAGCU ...........................................(((((.(((.....)))..(((((................)))))))))). ( -8.30 = -8.49 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:04 2006