
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,780,464 – 9,780,594 |
| Length | 130 |
| Max. P | 0.960480 |

| Location | 9,780,464 – 9,780,557 |
|---|---|
| Length | 93 |
| Sequences | 6 |
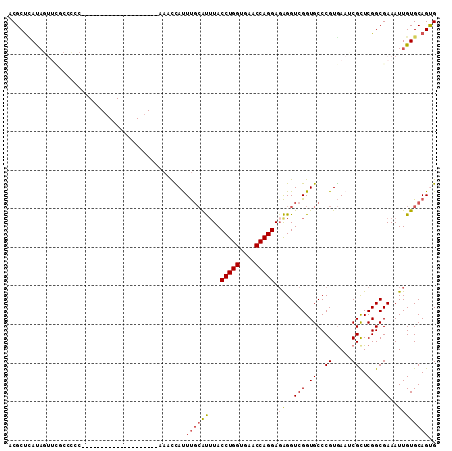
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9780464 93 - 23771897 ACGCUCAUAGUUUGCCCCCC-----A-------------AAUCCAUUUGCAUUUACCUGGUGAACCAGGAGAGGUCGGUGCCCGUGAAUCGCUCGGCGAAAUUGUGCAGUG .(((((((((((((((....-----.-------------................(((((....))))).(((..((((........)))))))))).)))))))).)))) ( -29.80) >DroSec_CAF1 75142 91 - 1 ACGCUCAUAGUUCGCCCCC--------------------AAACCAUUUGCAUUUACCUGGUGAACCAGGAGAGGUCGGUGCCCGUGAAUCGCUCGGCGAAAUUGUGCAGCG .(((((((((((((((...--------------------................(((((....))))).(((..((((........))))))))))).))))))).)))) ( -33.50) >DroSim_CAF1 77273 91 - 1 ACGCUCAUAGUUCGCCCCC--------------------AAACCAUUUGCAUUUACCUGGUGAACCAGGAUAGGUCGGUGCCCGUGAAUCGCUCGGCGAAAUUGUGCAGUG .(((((((((((((((...--------------------................(((((....)))))...(..((((........))))..))))).))))))).)))) ( -30.40) >DroEre_CAF1 77179 80 - 1 ACGCCUAUA------------------------------CAUCCACUUGCAU-UACCUGGUGAACCAGGAGAGGUCGGUGCCCGUGAAGCGCUCGGCGAAAUUGUGCAGUG .........------------------------------....((((.((((-..(((((....)))))....((((((((.......)))).))))......)))))))) ( -27.30) >DroYak_CAF1 86814 111 - 1 ACGCUGAUAUUUCACCACCCCUCCGAAAAGCAUUUUCACCAACUAUUUGCAUUUACCUGGUGAACCAGGAGAGGUCGGUGCCCGUGAAGCGUUCGGCGAAAUUGUGCAGUG .((((((..((((((((((((((......(((...............))).....(((((....))))).))))..))))...))))))...))))))............. ( -36.06) >DroAna_CAF1 80806 79 - 1 UCGCUUAC-------CCG-----------------------CUUACUC--GCUUACCUGGUGAACCAGGAGGGGUCGGUGCCCGUAAAACGCUCGGCGAAAUUGUGCAGCG .((((.((-------(((-----------------------(......--))...(((((....)))))..)))).)))).........((((..(((......))))))) ( -26.60) >consensus ACGCUCAUAGUUCGCCCCC____________________AAACCAUUUGCAUUUACCUGGUGAACCAGGAGAGGUCGGUGCCCGUGAAUCGCUCGGCGAAAUUGUGCAGUG ..............................................((((((...(((((....))))).((..(((.((..((.....))..)).)))..)))))))).. (-19.97 = -20.22 + 0.25)


| Location | 9,780,504 – 9,780,594 |
|---|---|
| Length | 90 |
| Sequences | 6 |
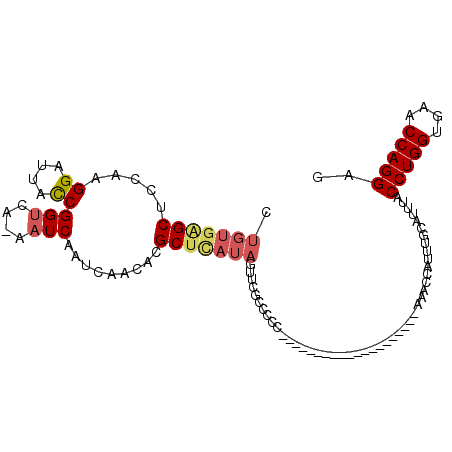
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9780504 90 - 23771897 CUGUGAGCUCCAAGGAUUACCGGUCA-AAUCAAUCAACACGCUCAUAGUUUGCCCCCC-----A-------------AAUCCAUUUGCAUUUACCUGGUGAACCAGGAG ((((((((.....((....))(((..-.....))).....))))))))..........-----.-------------................(((((....))))).. ( -22.10) >DroSec_CAF1 75182 88 - 1 CUGUGAGCUCCAAGGAUUACCGGUCA-AAUCAAUCAACACGCUCAUAGUUCGCCCCC--------------------AAACCAUUUGCAUUUACCUGGUGAACCAGGAG ((((((((.....((....))(((..-.....))).....)))))))).........--------------------................(((((....))))).. ( -22.10) >DroSim_CAF1 77313 88 - 1 CUGUGAGCUCCAAGGAUUACCGGUCA-AAUCAAUCAACACGCUCAUAGUUCGCCCCC--------------------AAACCAUUUGCAUUUACCUGGUGAACCAGGAU ((((((((.....((....))(((..-.....))).....)))))))).........--------------------................(((((....))))).. ( -22.10) >DroEre_CAF1 77219 77 - 1 CUGUGAGCUCCAAGGAUUACCGGCCA-ACUCAAUCAACACGCCUAUA------------------------------CAUCCACUUGCAU-UACCUGGUGAACCAGGAG .((..((......((((((..(((..-.............)))..))------------------------------.)))).))..)).-..(((((....))))).. ( -17.86) >DroYak_CAF1 86854 108 - 1 CUGUGAGCUCCAAGGAUUACCGGUCA-AAUCAAUCAACACGCUGAUAUUUCACCACCCCUCCGAAAAGCAUUUUCACCAACUAUUUGCAUUUACCUGGUGAACCAGGAG ..(((((((....(((.....(((.(-(((..((((......)))))))).))).....)))....)))....))))................(((((....))))).. ( -23.30) >DroAna_CAF1 80846 77 - 1 CUCGGGGCUCAAAGGAUUUUCGGUGGCAAUCAUGAAACUCGCUUAC-------CCG-----------------------CUUACUC--GCUUACCUGGUGAACCAGGAG ..((((.....(((((.(((((.((.....))))))).)).))).)-------)))-----------------------.......--.....(((((....))))).. ( -20.20) >consensus CUGUGAGCUCCAAGGAUUACCGGUCA_AAUCAAUCAACACGCUCAUAGUUCGCCCCC____________________AAACCAUUUGCAUUUACCUGGUGAACCAGGAG .(((((((.....((....))(((....))).........)))))))..............................................(((((....))))).. (-15.95 = -16.53 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:55:01 2006