
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 961,855 – 961,958 |
| Length | 103 |
| Max. P | 0.689698 |

| Location | 961,855 – 961,958 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -15.08 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 961855 103 + 23771897 GGAUUCGGUUUCGCCG------GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC-------GGCACAACAAUGUCCGUG-AAAUGCAAAACAAUAACAGUACUAACGAUAGCAUU-- ((((...(((..((((------((....))..((((((((......)))))))).)-------)))..)))...))))...-.(((((...((.......)).(....)...)))))-- ( -23.00) >DroPse_CAF1 6069 100 + 1 GUAA--CGUAACGUUC------GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC-------GGCAAAACAAUGUCCGUGCAAAUGCAAAACAAUAACAAUACUAACGACAGCA---- (.((--((...)))))------((((.(((..((((((((......))))))))..-------.)))......(((...(((....)))..)))................)))).---- ( -20.20) >DroWil_CAF1 6378 109 + 1 UGUU---GCUGCUCCU------GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUCCAGCUCUAGCACAACAAUGUCCGUG-AAAUGGAAAACAAUAACAAUAACAGCAACAGCAACAU ((((---((((....(------((((.(((..((((((((......))))))))((((..((....(((....)))....)-)..))))...........))).))))).)))))))). ( -30.50) >DroYak_CAF1 4682 109 + 1 AGAUUCGGUUUCGCUGCUGUACGCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC-------GGCACAACAAUGUCCGUG-AAAUGCAAAACAAUAACAGUACUAACGAUAGCAUU-- .......((((((.((((((.......(((((((((((((......)))))))).(-------((.(((....))))))..-..)))))........))))))....))).)))...-- ( -21.06) >DroAna_CAF1 4722 100 + 1 UGUUUCGGUUUCGUUC------GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC-------GGCAUAACAAUGUCCGUG-AAAUGCAAAACAAUAACAGAACUAACGAUAGC----- ((((...((((.((((------((........((((((((......))))))))..-------(((((....))))).)))-)...)).))))...))))..............----- ( -18.90) >DroPer_CAF1 5407 102 + 1 GUAA--CGUAACGUUC------GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC-------GGCAAAACAAUGUCCGUGCAAAUGCAAAACAAUAACAAUACUAACGACAGCAGC-- (.((--((...)))))------((((.(((..((((((((......))))))))..-------.)))......(((...(((....)))..)))................))))...-- ( -20.20) >consensus GGAU__GGUUUCGCUC______GCUGAUGUAUGCAUAAAAUAAUCCUUUUAUGCUC_______GGCACAACAAUGUCCGUG_AAAUGCAAAACAAUAACAAUACUAACGACAGCA____ ......................((((.(((..((((((((......)))))))).........((((......)))).............................))).))))..... (-15.08 = -14.72 + -0.36)

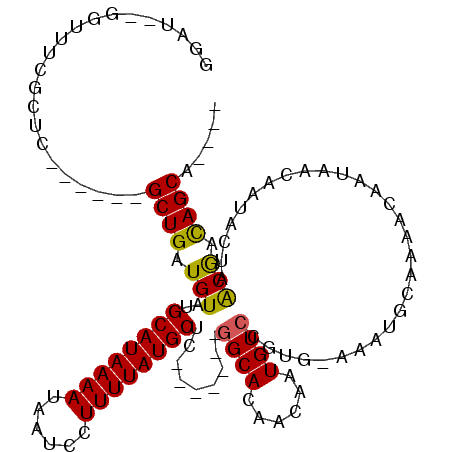
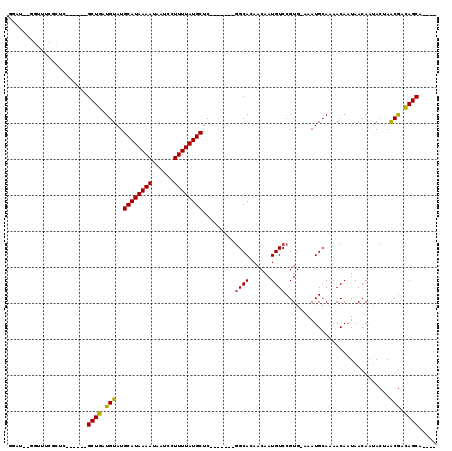
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:29 2006