| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,728,886 – 9,728,977 |
| Length | 91 |
| Max. P | 0.920417 |

| Location | 9,728,886 – 9,728,977 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
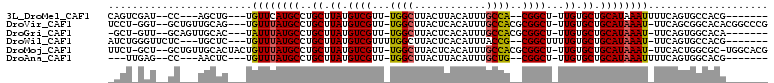
>3L_DroMel_CAF1 9728886 91 + 23771897 CAGUCGAU--CC---AGCUG---UGUUCAUGCCUGCUUAUGUCGUU-UGGCUUACUUACAUUUGCCA--CGGCU-UUGUGCUGCAUAAAUUUUCAGUGCCACG------- ........--..---.((((---.(((.((((..((....(((((.-.(((............))))--)))).-....)).)))).)))...))))......------- ( -20.30) >DroVir_CAF1 24725 101 + 1 UCCU-GGU--GCUGUUGCAG---UGUUUAUGCCUGCUUAUGUCGUU-UGGCUUACUCACAUUUGCCACGCGGCU-UUGUGCUGCAUAAAU-UUCAGCGGCACACGGCCCG ..((-(((--(((((((...---.((((((((..((....(((((.-((((............)))).))))).-....)).))))))))-..))))))))).))).... ( -40.10) >DroGri_CAF1 28774 93 + 1 -GCU-GUU--GCAGUUGCAC---UAUUUAUGCCUGCUUAUGUCGUU-UGGCUUACUCACAUUUGCCACGCGGCU-UUGUGCUGCAUAAAU-UUCAGUGGCACA------- -((.-...--)).((..(..---.((((((((..((....(((((.-((((............)))).))))).-....)).))))))))-....)..))...------- ( -29.80) >DroWil_CAF1 25152 94 + 1 AUCUGGGUUCUC---UGCUC---UGUUUAUGCCUGCUUAUGUCGUUUUGGCUUACUCACAUUUACCG--CGGCUUUUGUGCUGCAUAAAU-UUCAGUGCCACG------- .....(((...(---((...---.(((((((((.((.......))...)))...............(--((((......)))))))))))-..))).)))...------- ( -21.00) >DroMoj_CAF1 26127 103 + 1 UUCU-GCU--GCUGUUGCACUACUGUUUAUGCCUGCUUAUGUCGUU-UGGCUUACUCACAUUUGCCACGCGGCU-UUGUGCUGCAUAAAU-UUCACUGGCGC-UGGCACG ....-..(--(((..(((......((((((((..((....(((((.-((((............)))).))))).-....)).))))))))-.......))).-.)))).. ( -31.42) >DroAna_CAF1 22046 88 + 1 ---UUGAG--CC---AACUC---UGUUUAUGCCUGCUUAUGUCGUU-UGGCUUACUUACAUUUGCUG--CGGCU-UUGUGCUGCAUAAAUUUUCAGUGGCACG------- ---....(--((---(....---.((((((((..((....(((((.-.(((............))))--)))).-....)).))))))))......))))...------- ( -25.60) >consensus _UCU_GAU__GC___UGCAC___UGUUUAUGCCUGCUUAUGUCGUU_UGGCUUACUCACAUUUGCCA__CGGCU_UUGUGCUGCAUAAAU_UUCAGUGGCACG_______ ........................((((((((..((.((.((((...((((............))))..))))...)).)).)))))))).................... (-17.17 = -17.00 + -0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:46 2006