| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 957,499 – 957,603 |
| Length | 104 |
| Max. P | 0.999989 |

| Location | 957,499 – 957,603 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.61 |
| SVM decision value | 5.52 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
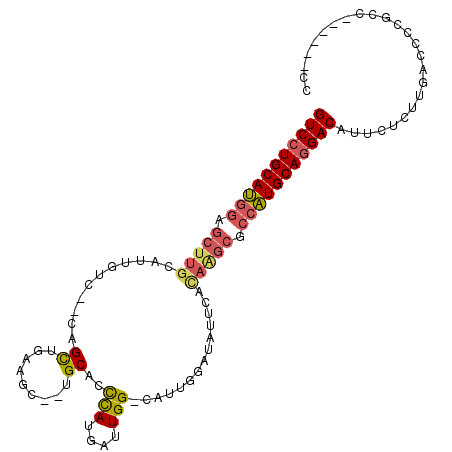
>3L_DroMel_CAF1 957499 104 + 23771897 GUCCUGCAUGGAGCUUGCAUUGUC--CUGCUCACGC--UGCACUCACGAUUGG-CAUUGGAUAUUCACAAGCGCCAUGCAAGACAUUCUCUUGACCCCCCCAGCCACAC (((.(((((((.(((((...((((--(((((..((.--((....))))...))-))..)))))....))))).))))))).)))......................... ( -32.40) >DroSec_CAF1 146 100 + 1 GUCCUGCAUGGAGCUUGCAUUAGUCGCAGCUGAAGC--UGCAUCCAUGAUUGG-CAUUGGAUAUUCAUAGGCGCCAUGCAGGACAUUCUCUUGACCCCGCC------CC (((((((((((.(((((........(((((....))--)))(((((((.....-)).))))).....))))).))))))))))).................------.. ( -42.30) >DroSim_CAF1 146 100 + 1 GUCCUGCAUGGAGCUUGCAUUGUCCGCAGCUGAAGC--UGCAUCCAUGAUUGG-CAUUGGAUAUUCACAAGCGCCAUGCAGGACAUGCUCUUGACCCCGCC------CC (((((((((((.(((((...((((((((((....))--)))..(((....)))-....)))))....))))).))))))))))).................------.. ( -47.40) >DroYak_CAF1 148 90 + 1 GUCCUGCAA-----------UGUC--CUGUGGCAGCAGUGCACCUAUGAUUGGGUAUACAGUUCCCACAAACGCCUUGCAGGACCCUCUGUUCACGUCAGU------CC (((((((((-----------.((.--.(((((.(((.(((.(((((....))))).))).))).)))))...)).)))))))))...(((.......))).------.. ( -33.70) >consensus GUCCUGCAUGGAGCUUGCAUUGUC__CAGCUGAAGC__UGCACCCAUGAUUGG_CAUUGGAUAUUCACAAGCGCCAUGCAGGACAUUCUCUUGACCCCGCC______CC (((((((((((.(((((...........((.........))..(((....)))..............))))).)))))))))))......................... (-23.74 = -24.55 + 0.81)

| Location | 957,499 – 957,603 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.24 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -24.51 |
| Energy contribution | -23.76 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
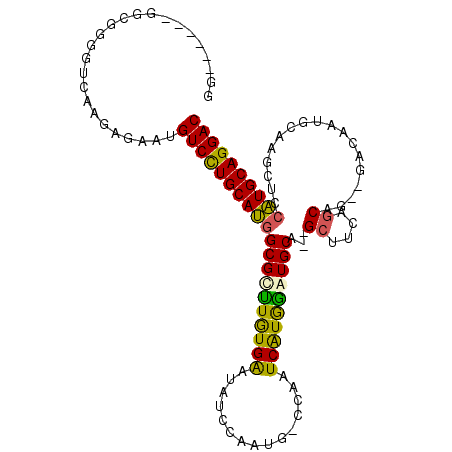
>3L_DroMel_CAF1 957499 104 - 23771897 GUGUGGCUGGGGGGGUCAAGAGAAUGUCUUGCAUGGCGCUUGUGAAUAUCCAAUG-CCAAUCGUGAGUGCA--GCGUGAGCAG--GACAAUGCAAGCUCCAUGCAGGAC ...(((((.....))))).......(((((((((((.((((((.....(((..((-(....(....).)))--((....)).)--))....)))))).))))))))))) ( -42.50) >DroSec_CAF1 146 100 - 1 GG------GGCGGGGUCAAGAGAAUGUCCUGCAUGGCGCCUAUGAAUAUCCAAUG-CCAAUCAUGGAUGCA--GCUUCAGCUGCGACUAAUGCAAGCUCCAUGCAGGAC ..------.................(((((((((((.((((((((..........-....)))))).((((--((....))))))..........)).))))))))))) ( -36.54) >DroSim_CAF1 146 100 - 1 GG------GGCGGGGUCAAGAGCAUGUCCUGCAUGGCGCUUGUGAAUAUCCAAUG-CCAAUCAUGGAUGCA--GCUUCAGCUGCGGACAAUGCAAGCUCCAUGCAGGAC ..------.((..........))..(((((((((((.((((((.....(((....-(((....)))..(((--((....))))))))....)))))).))))))))))) ( -44.90) >DroYak_CAF1 148 90 - 1 GG------ACUGACGUGAACAGAGGGUCCUGCAAGGCGUUUGUGGGAACUGUAUACCCAAUCAUAGGUGCACUGCUGCCACAG--GACA-----------UUGCAGGAC ..------.(((.......)))...(((((((((....(((((((.(.(.((.((((........)))).)).).).))))))--)...-----------))))))))) ( -30.30) >consensus GG______GGCGGGGUCAAGAGAAUGUCCUGCAUGGCGCUUGUGAAUAUCCAAUG_CCAAUCAUGGAUGCA__GCUUCAGCAG__GACAAUGCAAGCUCCAUGCAGGAC .........................((((((((((((((((((((...............))))))))))...((....))..................)))))))))) (-24.51 = -23.76 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:28 2006