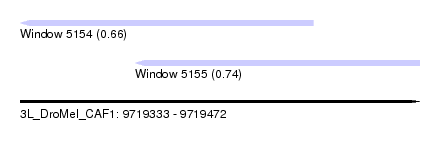
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,719,333 – 9,719,472 |
| Length | 139 |
| Max. P | 0.744767 |

| Location | 9,719,333 – 9,719,435 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9719333 102 - 23771897 CAUCU--AUGGGA----G-CUCC-------AGACUG--AGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCGGAGACUUGAUGGCGAGUGCCG .....--...((.----(-((((-------(((..(--(((....))))))))((((......(((((.(((((((((...))))))))).)))))(....)....)))))))).)). ( -34.60) >DroPse_CAF1 13238 105 - 1 UCUCUGCAGGGGACCUCG-CUCC-------GGUCUC--GGUCUGGGC---CUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCUGAGACUUGAUGGCGAGUGCCG ..........((..((((-(((.-------((((((--((((.....---..)))).......(((((.(((((((((...))))))))).))))).)))))).))..)))))..)). ( -39.60) >DroGri_CAF1 17252 111 - 1 ----GGGUACGGGUCUGGGGUCUAGUUGGGGGUCUAGUUGACUUGGU---CUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGCACUGAGACUUGAUGGCGAGUGCCG ----.(((((..((((.(((((((((..((((((..((((.(((((.---.((((((.....))))))...))))).)))).....))))))..))).)))))).).)))..))))). ( -35.80) >DroYak_CAF1 22851 102 - 1 CGUCU--AUGGGA----G-CUCC-------AGUCUG--GGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCGGAGACUUGAUGGCGAGUGCCG .....--...((.----(-((((-------.(((.(--((((((((((....))))......((((((.(((((((((...))))))))).)))))))))))))))).).)))).)). ( -36.20) >DroAna_CAF1 12413 101 - 1 UAUCU--GUGAGA----C-CUCC-------AGACUG--AGUCU-GGCUUUUCGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCAGAGACUGGAUGGCUGGUGCCG ..(((--(.((..----.-.)))-------)))((.--(((((-((((....)))))..((.((((((.(((((((((...))))))))).))))))))))))))..(((....))). ( -34.10) >DroPer_CAF1 13155 105 - 1 UCUCUGCAGGGGACCUCG-CUCC-------GGUCUC--GGUCUGGGC---CUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCUGAGACUUGAUGGCGAGUGCCG ..........((..((((-(((.-------((((((--((((.....---..)))).......(((((.(((((((((...))))))))).))))).)))))).))..)))))..)). ( -39.60) >consensus UAUCU__AUGGGA____G_CUCC_______AGUCUG__GGUCUUGGC___CUGGCCAAAUCGUGGUCGUAGUUAAGAUAACAUUUUGACUUUGGCCUGAGACUUGAUGGCGAGUGCCG ............................................(((...((.((((..((..(((((.(((((((((...))))))))).)))))...)).....)))).)).))). (-24.45 = -24.23 + -0.22)


| Location | 9,719,373 – 9,719,472 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.81 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -25.56 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9719373 99 - 23771897 UGUUAACUUUUUGUUCUCCGUGUGGCG---UUCCCGUCUCCAUCUAUGGGAGCUCCAGACUGAGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU ...............(((.((.(((.(---(((((((........)))))))).))).)).)))((((((((...((((((.....)))))).)))))))). ( -36.00) >DroSec_CAF1 14545 99 - 1 UGUUAACUUUUUGUUCUCCGUGUGGCG---UUCCCGUCUCCAUCUAUGCGAGCUCCAGACUGGGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU .................((((.(((.(---(((.(((........))).)))).))).)).))(((((((((...((((((.....)))))).))))))))) ( -28.40) >DroSim_CAF1 15055 99 - 1 UGUUAACUUUUUGUUCUCCGUGUGGCG---UUCCCGUCUCCAUCUAUGGGAGCUCCAGACUGGGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU .................((((.(((.(---(((((((........)))))))).))).)).))(((((((((...((((((.....)))))).))))))))) ( -35.50) >DroEre_CAF1 13758 99 - 1 UGCCAACUUGUUGUUCUCCGUGUGGCC---UUCCCGUCUCCAUCUAUGGGAGCUCCAGACUGGGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU .................((((.(((..---(((((((........)))))))..))).)).))(((((((((...((((((.....)))))).))))))))) ( -31.40) >DroYak_CAF1 22891 102 - 1 UGUUAACUUGUUGUUCGCCGUGUGGCCGGGUUCCCGUCUCCGUCUAUGGGAGCUCCAGUCUGGGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU ..((((((...((..(((.((.(((((((((((((((........)))))))).))((((........))))....))))).)).)))..)).))))))... ( -35.30) >consensus UGUUAACUUUUUGUUCUCCGUGUGGCG___UUCCCGUCUCCAUCUAUGGGAGCUCCAGACUGGGUCUUGGCUUUCUGGCCAAAUCGUGGUCGUAGUUAAGAU ...............(((.((.(((.....(((((((........)))))))..))).)).)))((((((((...((((((.....)))))).)))))))). (-25.56 = -26.04 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:39 2006