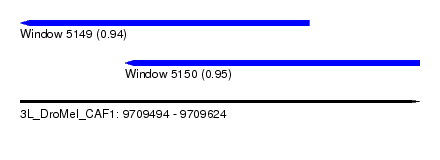
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,709,494 – 9,709,624 |
| Length | 130 |
| Max. P | 0.945840 |

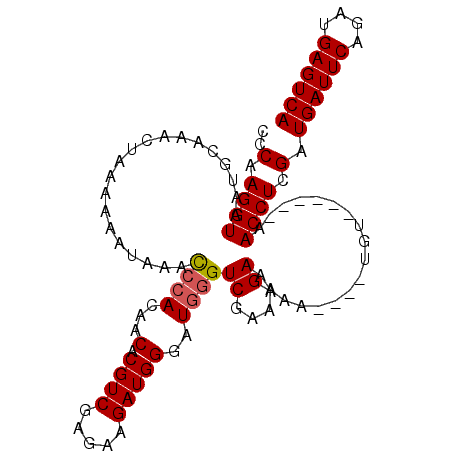
| Location | 9,709,494 – 9,709,588 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9709494 94 - 23771897 ACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAA----UGU------AACCUCGAUGAUUCAGAUGAGUCAUGAAUCAUUAAGAAUAAUGAAUCAUAAGUGAACA ..((((((......(.((...((....))...)----).)------...))))))(((((....)))))((((.((((((....)))))).))))......... ( -21.60) >DroSec_CAF1 4657 94 - 1 ACGUCGAGAAGAUGGGUUGGGUCGAAAGAAAAA----UGU------AACCUCGAUGAUUCAGAUGAGUCAUGAAUCAUAAUGAAUCAUGAAUCAUAAGUAAGCA .(((((((....((.(((...((....))..))----).)------)..)))))))......((((.((((((.((.....)).)))))).))))......... ( -25.70) >DroEre_CAF1 4194 92 - 1 ACGUCGAGAAGAUGGGAUGGGUCGAAAGA----UACUUGUAU----AACCUCGAUGAUUCAGUUGAGUCAUGAAUCAUAAGCAUUCUU-AG---CGCGUGAGCA .(((((((...(((.((.(..((....))----..))).)))----...))))))).....((((((..(((.........))).)))-))---)((....)). ( -26.70) >DroYak_CAF1 13163 100 - 1 ACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAAUAUAUGUAUAUACAACCUCGAUGAUUCAGAUGAGUCAUGAAUCAUAAGCAUUCUU-AG---CCAAUGAGCA .((((.....))))(((((((((....))........(((....))).))...((((((((((....)).))))))))...)))))..-.(---(......)). ( -20.30) >consensus ACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAA____UGU______AACCUCGAUGAUUCAGAUGAGUCAUGAAUCAUAAGCAAUCAU_AA___CAAGUGAGCA .((((.....))))..((((.((....)).....................((.(((((((....))))))))).)))).......................... (-13.55 = -13.80 + 0.25)

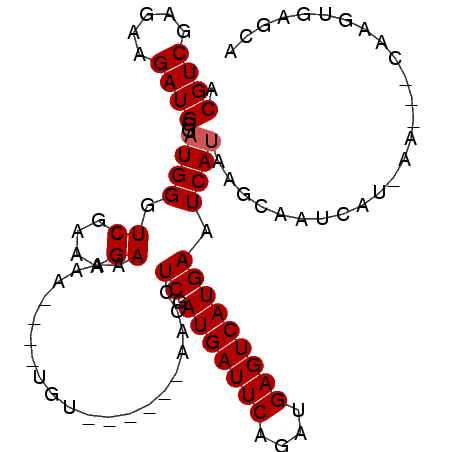
| Location | 9,709,528 – 9,709,624 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9709528 96 - 23771897 CCCAAGGUAAUGCAAACUUAAAAAAUAACCCACAACACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAA----UGU------AACCUCGAUGAUUCAGAUGAGUCA ..(.((((..((((..............((((...(.((((.....)))))..))))((....))....----)))------))))).).((((((....)))))) ( -22.30) >DroSec_CAF1 4691 96 - 1 CCCAAGGUAAUGCAAACUAAAAAAUAAAUAAACCACACGUCGAGAAGAUGGGUUGGGUCGAAAGAAAAA----UGU------AACCUCGAUGAUUCAGAUGAGUCA ..(.((((..((((..................(((..((((.....))))...))).((....))....----)))------))))).).((((((....)))))) ( -20.20) >DroEre_CAF1 4224 97 - 1 CCCAAGGUAAUGCAAACUAAAAAAUA-ACCCACAACACGUCGAGAAGAUGGGAUGGGUCGAAAGA----UACUUGUAU----AACCUCGAUGAUUCAGUUGAGUCA ..(.((((.((((((...........-.((((...(.((((.....)))))..))))((....))----...))))))----.)))).).((((((....)))))) ( -26.00) >DroYak_CAF1 13193 106 - 1 CCCAAGGUAAUGCAAACUAAAAAAUAAACCCACAACACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAAUAUAUGUAUAUACAACCUCGAUGAUUCAGAUGAGUCA ..(.((((.(((((..............((((...(.((((.....)))))..))))((....))........))))).....)))).).((((((....)))))) ( -21.80) >consensus CCCAAGGUAAUGCAAACUAAAAAAUAAACCCACAACACGUCGAGAAGAUGGGAUGGGUCGAAAGAAAAA____UGU______AACCUCGAUGAUUCAGAUGAGUCA ..(.((((....................((((...(.((((.....)))))..))))((....))..................)))).).((((((....)))))) (-17.54 = -17.85 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:34 2006