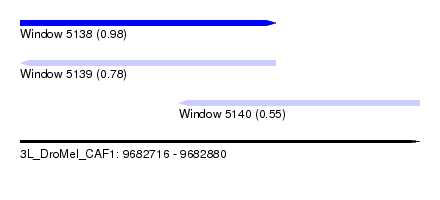
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,682,716 – 9,682,880 |
| Length | 164 |
| Max. P | 0.983407 |

| Location | 9,682,716 – 9,682,821 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -30.81 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
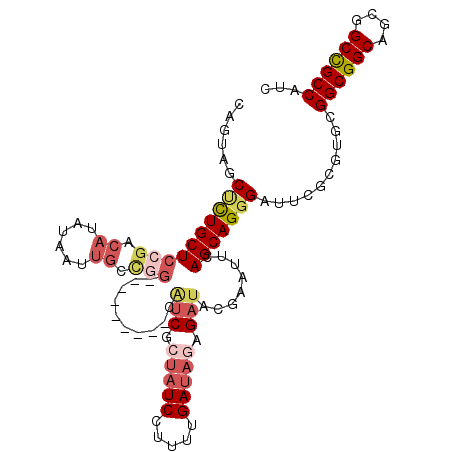
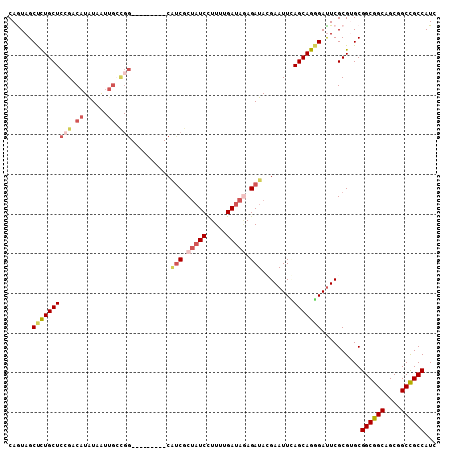
>3L_DroMel_CAF1 9682716 105 + 23771897 AAUAGUAGCAAUAAAUUUUGUUAUCCAACGGCAGCGGCCAUGGAGAAUAUGUUCCAGAACAAUCAGAUGGCGGCCGCUGCCGCCGCACGCGAAUCCCUGCUGAAU .....(((((......(((((.......(((((((((((.(((((......)))))((....)).......)))))))))))......)))))....)))))... ( -36.42) >DroSec_CAF1 4419 105 + 1 AAUAGUAGCAAUAAAUUUUGUUAUCCAACGGCGGCGGCCAUGGAGAAUAUGUUCCAGAACAAUCACAUGGCAGCCGCUGCCGCCGCACGCGAAUCCCUGCUGAAU ..((((((....................(((((((.((((((((.....((((....)))).)).)))))).))))))).(((.....))).....))))))... ( -34.80) >DroSim_CAF1 1661 105 + 1 AAUAGUAGCAAUAAAUUUUGUUAUCCAACGGCGGCGGCCAUGGAGAAUAUGUUCCAGAACAAUCAGAUGGCAGCCGCUGCCGCCGCACGCGAAUCCCUGCUGAAU ..((((((....................(((((((.(((((((((......)))).((....))..))))).))))))).(((.....))).....))))))... ( -32.20) >DroEre_CAF1 4194 105 + 1 AAUAGUAGCAAUAAAUUUUGUUAUCCUACGGCGGCAGCCAUGGAGAAUAUGUUCCAAAACAAUCAGAUGGCGGCCGCUGCCGCCGCACGCGAAUCCCUGCUGAAU ....(((((((......)))))))....((((((((((..(((((......)))))............((...))))))))))))((.(((......)))))... ( -32.40) >DroYak_CAF1 1671 105 + 1 AAUAGUAGCAAUAAAUUUUGUUAUCCUACGGCGGCGGCCAUGGAGAAUAUGUUCCAAAACAAUCAAAUGGCGGCCGCUGCCGCCGCACGCGAAUCCCUGCUAAAU .....(((((......(((((.......(((((((((((.(((((......)))))...((......))..)))))))))))......)))))....)))))... ( -35.82) >DroAna_CAF1 4150 105 + 1 AAUAGCAGCAACAAGUAUUGUUAUCCCACGGCGGCGGCCAUGGAGAACAUGUUCCAGAGCAAUCAGAUGGCAGCCGCCGCCGCCGCUCGCGAAUCGCUGCUCAAU ...((((((((((.....))))......(((((((.(((((((((......)))).((....))..))))).))))))).(((.....)))....)))))).... ( -41.60) >consensus AAUAGUAGCAAUAAAUUUUGUUAUCCAACGGCGGCGGCCAUGGAGAAUAUGUUCCAGAACAAUCAGAUGGCAGCCGCUGCCGCCGCACGCGAAUCCCUGCUGAAU ..((((((....................(((((((.(((((.((.....((((....)))).))..))))).))))))).(((.....))).....))))))... (-30.81 = -30.62 + -0.19)


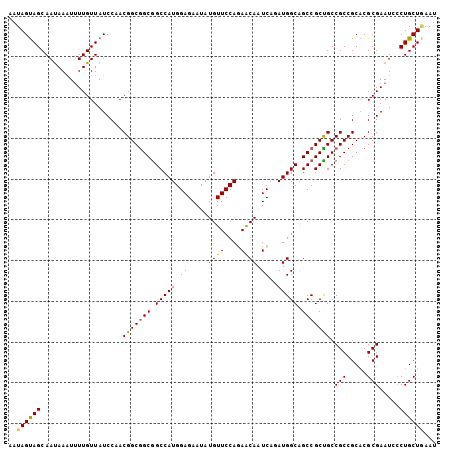
| Location | 9,682,716 – 9,682,821 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.65 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9682716 105 - 23771897 AUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUCUGAUUGUUCUGGAACAUAUUCUCCAUGGCCGCUGCCGUUGGAUAACAAAAUUUAUUGCUACUAUU ....((((((((((...((.(((((((((((((((......((....))((((........))))))))))))))))))).)).....))))).)))))...... ( -39.50) >DroSec_CAF1 4419 105 - 1 AUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCUGCCAUGUGAUUGUUCUGGAACAUAUUCUCCAUGGCCGCCGCCGUUGGAUAACAAAAUUUAUUGCUACUAUU ....((((((((((...((.(((((((...(((.((((((.((.((((....))))...))..)))))).)))))))))).)).....))))).)))))...... ( -34.30) >DroSim_CAF1 1661 105 - 1 AUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCUGCCAUCUGAUUGUUCUGGAACAUAUUCUCCAUGGCCGCCGCCGUUGGAUAACAAAAUUUAUUGCUACUAUU ....((((((((((...((.(((((((.((((((.......((....))((((........)))).)))))).))))))).)).....))))).)))))...... ( -32.40) >DroEre_CAF1 4194 105 - 1 AUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUCUGAUUGUUUUGGAACAUAUUCUCCAUGGCUGCCGCCGUAGGAUAACAAAAUUUAUUGCUACUAUU ....((((((((((..(.((((((((((((((...))............((((........))))..)))))))))))).).......))))).)))))...... ( -34.70) >DroYak_CAF1 1671 105 - 1 AUUUAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUUUGAUUGUUUUGGAACAUAUUCUCCAUGGCCGCCGCCGUAGGAUAACAAAAUUUAUUGCUACUAUU ...(((((((((((..(.(((((((((...(((((.((......))...((((........)))))))))))))))))).).......))))).))))))..... ( -34.00) >DroAna_CAF1 4150 105 - 1 AUUGAGCAGCGAUUCGCGAGCGGCGGCGGCGGCUGCCAUCUGAUUGCUCUGGAACAUGUUCUCCAUGGCCGCCGCCGUGGGAUAACAAUACUUGUUGCUGCUAUU ....((((((..((((((.(((((((((((....)))............((((........))))..))))))))))))))..((((.....))))))))))... ( -45.30) >consensus AUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUCUGAUUGUUCUGGAACAUAUUCUCCAUGGCCGCCGCCGUUGGAUAACAAAAUUUAUUGCUACUAUU ....(((((..(((((((.((((((((.((....)).............((((........))))..))))))))))).))))...........)))))...... (-30.93 = -30.65 + -0.28)

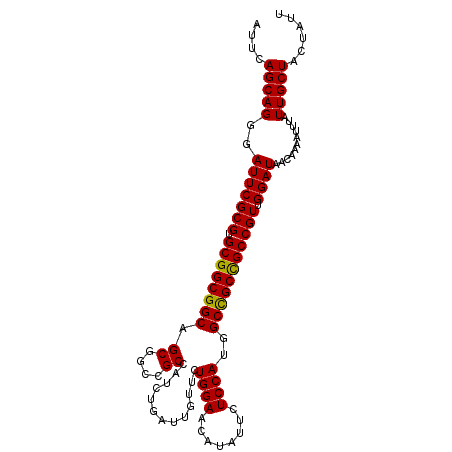
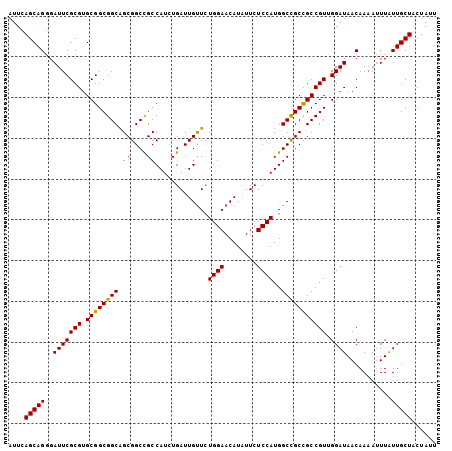
| Location | 9,682,781 – 9,682,880 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -24.70 |
| Energy contribution | -26.12 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9682781 99 - 23771897 CAGUAGCUCUGCUCCGCCAUAUAAUUGCCGG---------CAUCGCUAUCCUUUUGAUAAAGAUAUGAAUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUC .....((.((((((((((.((....))...(---------((.(((.((((((((((.............)))).))))))..))))))))))).)))))..)).... ( -31.72) >DroPse_CAF1 4740 108 - 1 CAGUAGCCUUGCUCAGACAUGUAGUUGGCGGCUGCCUCUGCGCCGCUCUCCUUCUGAUAGAGAUACGAAUUGAGCAGGGACUCGCGGGCGGCGGCGGCUGCCGCCAUC ..((((((((((((((...((((....(((((.((....)))))))((((.........))))))))..)))))))))).)).)).((((((((...))))))))... ( -47.10) >DroSim_CAF1 1726 99 - 1 CAGUAGCUCUGCUCCGUCAUAUAAUUGCCGG---------CAUCGCUAUCCUUUUGAUAGAGAUAGGAAUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCUGCCAUC ..((((((((((((((.(((...........---------.(((.(((((.....))))).)))..((((((.....))))))..)))))).))))).)))))).... ( -32.80) >DroEre_CAF1 4259 99 - 1 CAAUAGCUCUGCUCCGACAUGUAAUUGCUGG---------CAUCGCUAUCUUUUUGAUAGAGAUAGGAAUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUC ..........((.((((...((..(((((((---------..((.((((((((.....))))))))))..)))))))..))))).).))((((((....))))))... ( -36.70) >DroYak_CAF1 1736 99 - 1 CAAUAGCUCUGCUCCGUCAUGUAGUUGCUGG---------CAUCGCUAUCUUUUUGAUAGAGAUAUGAAUUUAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUU ...((((.((((........))))..))))(---------((.((((((((((.....))))))).((((((.....))))))))))))((((((....))))))... ( -36.90) >DroAna_CAF1 4215 90 - 1 CAGUAGCCCUGCUCCGACAUAUAA------------------UCUCCAUCCUUCUGAUACAGAUACGAAUUGAGCAGCGAUUCGCGAGCGGCGGCGGCGGCUGCCAUC ..((((((((((((((.(.....(------------------(((..(((.....)))..)))).(((((((.....)))))))...)))).))))).)))))).... ( -30.20) >consensus CAGUAGCUCUGCUCCGACAUAUAAUUGCCGG_________CAUCGCUAUCCUUUUGAUAGAGAUACGAAUUCAGCAGGGAUUCGCGUGCGGCGGCAGCGGCCGCCAUC ......((((((((((.((......)).)))..........(((.(((((.....))))).)))........)))))))..........((((((....))))))... (-24.70 = -26.12 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:24 2006