
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,677,471 – 9,677,606 |
| Length | 135 |
| Max. P | 0.956650 |

| Location | 9,677,471 – 9,677,574 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -20.53 |
| Energy contribution | -22.95 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9677471 103 + 23771897 GAGGGAGCCGCUGGUGUCUCCUCCGGCUGUCCAGCAGCCG------AGGCUCCCUCGUCGGAGCCGGAUUUCUGCAUCCUGGCCAAUGCAAAAAUCAAAGGAUAUUGGC (((((((((...((.....))..((((((.....))))))------.)))))))))(((((..((.(((((.(((((........))))).)))))...))...))))) ( -47.00) >DroPse_CAF1 76736 109 + 1 GCCUGCUGUGACGGCGGCUCCUCUGGCUGGGCGGAUGCCGCCCCAGCAGCUGCCUCGUCGGAGCUUCCUUUUUGCAUCCUCGCAGAUGCACAAAUCGAUUUGCAUUGUG ((..(((.(((((((((((......(((((((((...))).)))))))))))))..)))).))).((..(((((((((......)))))).)))..))...))...... ( -43.40) >DroSim_CAF1 76685 103 + 1 GAGGGAGCCGCUGGCGUCUCCUCAGGCUGGCCAGCAGCCG------AAGCUCCCUCGUCGGAGCCGGAUUUCUGCAUCCUGCCCAAUGCACAAAUCAAAGGAUAUUGGU ((((((((.((((((((((....))))..)))))).....------..))))))))((.(((..(((....)))..))).))((((((..(........)..)))))). ( -39.40) >DroEre_CAF1 73585 103 + 1 GAGGGAGCCGCUGGCGUCUCCUCCGGCUGGCCAGCAGCCG------AGGCUCCCUCAUCGGAGCCAGAUUUCUGCAUCCUGGCCAAUGCACAAAUCAACACAAAUUGGC ((((((((((((((.(....).)))))((((.....))))------.)))))))))...(((..(((....)))..)))..((((((................)))))) ( -43.79) >DroYak_CAF1 73796 103 + 1 GAGGAAGCAGCUGGCGUCUCCUCCGUCUGGCCAGCAGCCG------AGGCUCCUUCAUCGGAGCCCGAUUUCUGCAUCCUGGCCAAUGCACAAAUCAACGCAAAUUGCA (((((.((.....))...))))).((.((((((((((.((------.((((((......))))))))....))))....))))))..))..........((.....)). ( -37.10) >DroPer_CAF1 75715 109 + 1 GCCUGCUGUGACGGCGGCUCCUCUGGCUGGGCGGAUGCCGCCCCAGCAGCUGCCUCGUCGGAGCUUCCUUUUUGCAUCCUCGCAGAUGCACAAAUCGAUUUGCAUUGUG ((..(((.(((((((((((......(((((((((...))).)))))))))))))..)))).))).((..(((((((((......)))))).)))..))...))...... ( -43.40) >consensus GAGGGAGCCGCUGGCGUCUCCUCCGGCUGGCCAGCAGCCG______AAGCUCCCUCGUCGGAGCCGGAUUUCUGCAUCCUGGCCAAUGCACAAAUCAAAGCAAAUUGGC (((((.((.......)).))))).(((((.....)))))........((((((......)))))).(((((.(((((........))))).)))))............. (-20.53 = -22.95 + 2.42)
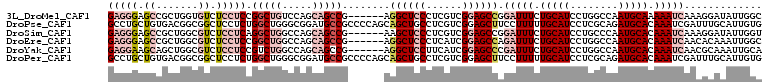

| Location | 9,677,471 – 9,677,574 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -44.53 |
| Consensus MFE | -24.07 |
| Energy contribution | -26.27 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9677471 103 - 23771897 GCCAAUAUCCUUUGAUUUUUGCAUUGGCCAGGAUGCAGAAAUCCGGCUCCGACGAGGGAGCCU------CGGCUGCUGGACAGCCGGAGGAGACACCAGCGGCUCCCUC (((((......))((((((((((((......)))))))))))).)))......(((((((((.------((((((.....)))))((.(....).)).).))))))))) ( -48.30) >DroPse_CAF1 76736 109 - 1 CACAAUGCAAAUCGAUUUGUGCAUCUGCGAGGAUGCAAAAAGGAAGCUCCGACGAGGCAGCUGCUGGGGCGGCAUCCGCCCAGCCAGAGGAGCCGCCGUCACAGCAGGC .....(((...(((.(((.(((((((....))))))).)))((.....))..))).))).((((((.((((((.(((...........)))))))))....)))))).. ( -44.00) >DroSim_CAF1 76685 103 - 1 ACCAAUAUCCUUUGAUUUGUGCAUUGGGCAGGAUGCAGAAAUCCGGCUCCGACGAGGGAGCUU------CGGCUGCUGGCCAGCCUGAGGAGACGCCAGCGGCUCCCUC .............(((((.((((((......)))))).))))).((((((......)))))).------.((((((((((...((...))....))))))))))..... ( -40.00) >DroEre_CAF1 73585 103 - 1 GCCAAUUUGUGUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCUGGCUCCGAUGAGGGAGCCU------CGGCUGCUGGCCAGCCGGAGGAGACGCCAGCGGCUCCCUC ((((.........(((((.((((((......)))))).)))))))))......(((((((((.------((((((.....)))))((.(....).)).).))))))))) ( -48.10) >DroYak_CAF1 73796 103 - 1 UGCAAUUUGCGUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCGGGCUCCGAUGAAGGAGCCU------CGGCUGCUGGCCAGACGGAGGAGACGCCAGCUGCUUCCUC .(((.((.(((((..(((((...(((((((....((((...(((((((((......)))))).------)))))))))))))))))))...))))).)).)))...... ( -42.80) >DroPer_CAF1 75715 109 - 1 CACAAUGCAAAUCGAUUUGUGCAUCUGCGAGGAUGCAAAAAGGAAGCUCCGACGAGGCAGCUGCUGGGGCGGCAUCCGCCCAGCCAGAGGAGCCGCCGUCACAGCAGGC .....(((...(((.(((.(((((((....))))))).)))((.....))..))).))).((((((.((((((.(((...........)))))))))....)))))).. ( -44.00) >consensus CCCAAUAUACAUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCAGGCUCCGACGAGGGAGCCU______CGGCUGCUGGCCAGCCGGAGGAGACGCCAGCGGCUCCCUC .............(((((.((((((......)))))).))))).((((((......))))))........(((.((((((...((...))....)))))).)))..... (-24.07 = -26.27 + 2.20)

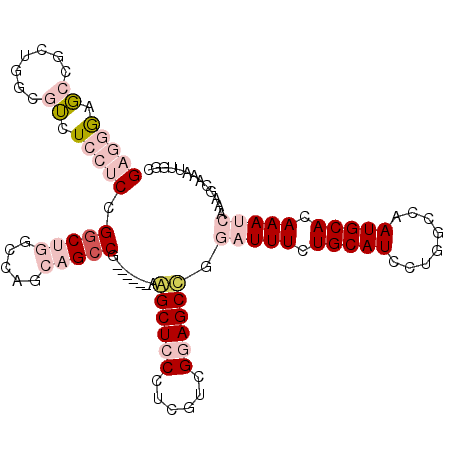
| Location | 9,677,502 – 9,677,606 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.33 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9677502 104 - 23771897 ACUACGAGCAACGAAACAAUCAAUCAAUA-AUGGCCAAUAUCCUUUGAUUUUUGCAUUGGCCAGGAUGCAGAAAUCCGGCUCCGACGAGGGAGCCU------CGGCUGCUG ......((((.(((...............-.(((((((((((....)))......))))))))((((......))))((((((......)))))))------))..)))). ( -34.00) >DroPse_CAF1 76767 105 - 1 A----UAGCAACCGAAAAAUCAAUCAAUA--UGCACAAUGCAAAUCGAUUUGUGCAUCUGCGAGGAUGCAAAAAGGAAGCUCCGACGAGGCAGCUGCUGGGGCGGCAUCCG .----..(((...((....)).......(--(((((((...........)))))))).)))..((((((.....((.((((((.....)).)))).))......)))))). ( -30.20) >DroSim_CAF1 76716 104 - 1 ACUACGAGCAACGAAACAAUAAAUCAAUA-AUGACCAAUAUCCUUUGAUUUGUGCAUUGGGCAGGAUGCAGAAAUCCGGCUCCGACGAGGGAGCUU------CGGCUGCUG ......((((.((((..............-................(((((.((((((......)))))).)))))..(((((......)))))))------))..)))). ( -28.30) >DroEre_CAF1 73616 105 - 1 GCCAAGAGAAACGAAACAAUCAAUCAAUAAAUGGCCAAUUUGUGUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCUGGCUCCGAUGAGGGAGCCU------CGGCUGCUG (((............................(((((((..((..(......)..)))))))))((((......))))((((((......)))))).------.)))..... ( -31.80) >DroYak_CAF1 73827 104 - 1 GCCAAGAGAAACGAAACAAUCAAUCAAUA-AUGUGCAAUUUGCGUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCGGGCUCCGAUGAAGGAGCCU------CGGCUGCUG (((.........................(-((((.......)))))(((((.((((((......)))))).)))))(((((((......)))))))------.)))..... ( -31.90) >DroPer_CAF1 75746 105 - 1 A----UAGCAACCGAAAAAUCAAUCAAUA--UGCACAAUGCAAAUCGAUUUGUGCAUCUGCGAGGAUGCAAAAAGGAAGCUCCGACGAGGCAGCUGCUGGGGCGGCAUCCG .----..(((...((....)).......(--(((((((...........)))))))).)))..((((((.....((.((((((.....)).)))).))......)))))). ( -30.20) >consensus AC_A_GAGCAACGAAACAAUCAAUCAAUA_AUGCCCAAUAUACAUUGAUUUGUGCAUUGGCCAGGAUGCAGAAAUCAGGCUCCGACGAGGGAGCCU______CGGCUGCUG ..............................................(((((.((((((......)))))).))))).((((((......))))))................ (-18.63 = -18.93 + 0.31)

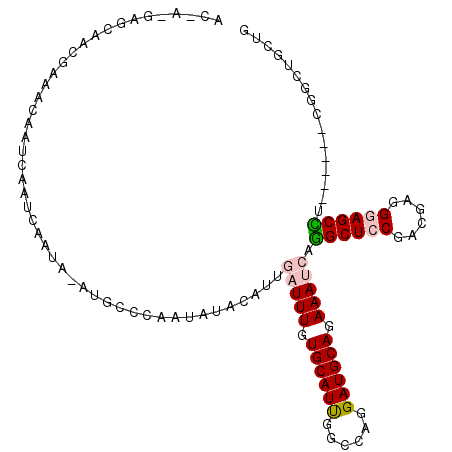

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:14 2006