
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,660,343 – 9,660,447 |
| Length | 104 |
| Max. P | 0.648450 |

| Location | 9,660,343 – 9,660,447 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -23.99 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
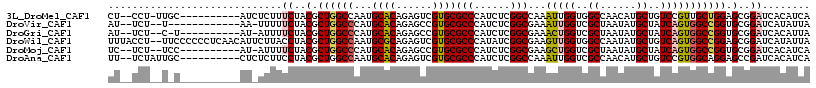
>3L_DroMel_CAF1 9660343 104 - 23771897 CU--CCU-UUGC----------AUCUCUUUCUACGCUGGCCAAUGCACAGAGUCGUGCGCCCAUCUCGGCCAAAUUGGUGGCCAACAUGCUGUCCGUUGCUGGAGCGGAUCACAUCA ..--...-....----------............(.((((((..((((......))))(((......)))........)))))).)(((..(((((((.....)))))))..))).. ( -29.00) >DroVir_CAF1 57422 100 - 1 AU--UCU--U------------AA-UUUUUCUACGCUGGCCCAUGCACAGAGCCGUGCGCCCAUCUCGGCGAAAUUGGUCGCUAAUAUGCUAUCAGUGGCCGGUGCGGAUCAUAUUA ..--...--.------------..-....((..((((((((...((((......))))(((......)))...((((((.((......)).))))))))))))))..))........ ( -33.60) >DroGri_CAF1 52759 101 - 1 AU--UCU--C-U----------AU-AUUUUCUACGCUGGCCCAUGCACAGAGCCGUGCGCCCAUCUCGGCGAAACUGGUCGCUAAUAUGCUAUCAGUGGCCGGUGCGGAUCACAUUA ..--...--.-.----------..-....((..((((((((...((((......))))(((......)))...((((((.((......)).))))))))))))))..))........ ( -35.90) >DroWil_CAF1 77855 115 - 1 UUUACCU--UUCCCCCCUCAACAUUCUUACCUACGCUGGCCAAUGCGCAGAGUCGUGCGCCCAUAUCGGCGAAGUUGGUGGCCAAUAUGCUGUCAGUGGCCGGAGCGGAUCAUAUUA ....((.--((((...............(((..((((((.....(((((......))))).....)))))).....)))(((((...((....)).))))))))).))......... ( -32.10) >DroMoj_CAF1 59404 102 - 1 UC--UCU--UCC----------AU-AUUUUCUACGCUGGCCCAUGCACAGAGCCGUGCGCCCAUCUCGGCGAAGCUGGUCGCUAAUAUGCUAUCAGUGGCCGGUGCGGAUCACAUCA ..--...--...----------..-....((..((((((((...((((......))))(((......)))...((((((.((......)).))))))))))))))..))........ ( -36.20) >DroAna_CAF1 67869 105 - 1 UU--UCUAUUGC----------CUCUCUUCCUACGCUGGCCAAUGCACAGAGUCGUGCGCCCAUCUCGGCCAAAUUGGUCGCCAACAUGCUGUCCGUGGCAGGAGCCGAUCACAUCA ..--...((((.----------..(((((.(((((..(((((((((((......))))(((......)))...)))))))((......))....))))).))))).))))....... ( -28.90) >consensus UU__UCU__U_C__________AU_UUUUUCUACGCUGGCCAAUGCACAGAGCCGUGCGCCCAUCUCGGCGAAAUUGGUCGCCAAUAUGCUAUCAGUGGCCGGAGCGGAUCACAUCA .............................((..(.((((((...((((......))))(((......)))...(((((..((......))..))))))))))).)..))........ (-23.99 = -24.30 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:09 2006