
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,649,900 – 9,650,034 |
| Length | 134 |
| Max. P | 0.704244 |

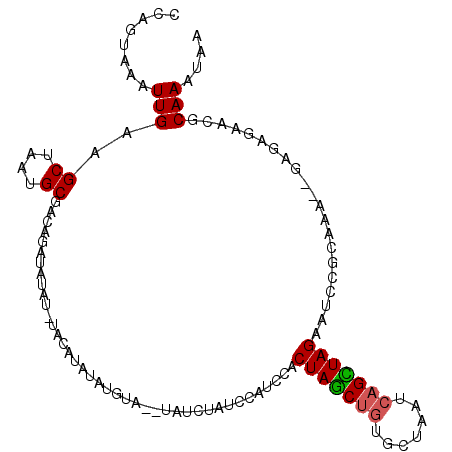
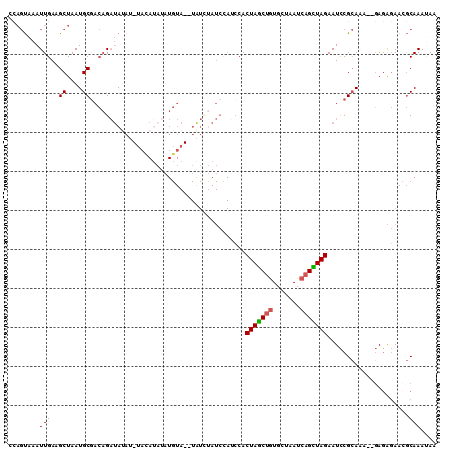
| Location | 9,649,900 – 9,650,002 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -10.63 |
| Energy contribution | -10.60 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
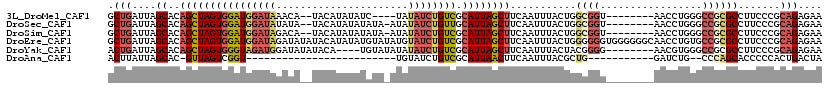
>3L_DroMel_CAF1 9649900 102 - 23771897 CCAGUAAAUUGAAGCUAAUGCGACAGAUAUA----GAUAUAUGUA--UGUUUAUCCAUCCACUAGCUGUGCUAAUCAGCUAGAAUCCGCAAA--GAGAGAAGGCAAAGAG (((((........)))..((((...((((((----(((((....)--)))))))..)))..(((((((.......)))))))....))))..--.......))....... ( -20.00) >DroSec_CAF1 45441 105 - 1 CCAGUAAAUUGAAGCUAAUGCAACAGAUAUAU-UAUAUAUAUGUA--UAUAUAUCCAUCCACUAGCUGUGCUAAUCAGCUAGAAUCCGCAAA--GAGAGAACGCAAAUAG ........(((...((..(((....(((((((-.((((....)))--))))))))......(((((((.......))))))).....)))..--.))......))).... ( -20.10) >DroSim_CAF1 48045 105 - 1 CCAGUAAAUUGAAGCUAAUGCGACAGAUAUAU-UAUAUAUAUGUA--UGUCUAUCCAUCCACUAGCUGUGCUAAUCAGCUAGAAUCCGCAAA--GAGAGAACGCAAAUAG ........(((...((..(((((((.((((((-.....)))))).--))))..........(((((((.......))))))).....)))..--.))......))).... ( -22.20) >DroEre_CAF1 45355 110 - 1 CCAGUAAAUUGAAGCUAAUGCGACAGAUACAUAUACAUAUAUGUAUAUAUCUAUCCAUCCACUAGCUGUGCUAAUCAGCUAGAAUCCGCACAGGGAGAGAACGCAAAUAA ..................((((....((((((((....))))))))...(((.(((.....(((((((.......)))))))....(.....)))).))).))))..... ( -25.90) >DroYak_CAF1 45651 106 - 1 GUAGUAAAUUGAAGCUAAUGCGACAGAUAUAUAUACA----UGUAUAUAUCCAUCUACCCACUAGCUGUGCUAAUCAGUUAGAAUCCGCAAGAGGAGAGAACGCAAAUAA .((((........)))).((((...(((((((((...----.)))))))))..(((..((.(((((((.......)))))))....(....).))..))).))))..... ( -27.50) >DroPer_CAF1 48041 82 - 1 CCCCCUAAUUGAAGCUAAUGCGAAUGCCAUA--------------------------AGCACUAACAAUGCUAAUCAGUUAGAAUUCGAAUCCGUAAGCA--ACAAACAA ........(((..(((.((((((((((....--------------------------.)).(((((...........))))).)))))....))).))).--.))).... ( -12.30) >consensus CCAGUAAAUUGAAGCUAAUGCGACAGAUAUAU_UACAUAUAUGUA__UAUCUAUCCAUCCACUAGCUGUGCUAAUCAGCUAGAAUCCGCAAA__GAGAGAACGCAAAUAA ........(((..((....))........................................(((((((.......))))))).....................))).... (-10.63 = -10.60 + -0.03)

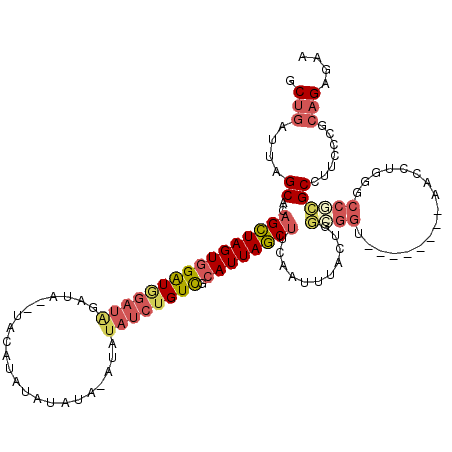
| Location | 9,649,929 – 9,650,034 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.89 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.55 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9649929 105 + 23771897 GCUGAUUAGCACAGCUAGUGGAUGGAUAAACA--UACAUAUAUC----UAUAUCUGUCGCAUUAGCUUCAAUUUACUGGCGGU--------AACCUGGGCCGCGCCUUCCCGCAGAGAA ((.((...((..((((((((((((((((.(.(--((....))).----).)))))))).))))))))...........(((((--------.......)))))))..))..))...... ( -28.10) >DroSec_CAF1 45470 108 + 1 GCUGAUUAGCACAGCUAGUGGAUGGAUAUAUA--UACAUAUAUAUA-AUAUAUCUGUUGCAUUAGCUUCAAUUUACUGGCGGU--------AACCUGGGCCGCGCCUUCCCGCAGAGAA ((.((...((..((((((((((((((((((((--((......))).-))))))))))).))))))))...........(((((--------.......)))))))..))..))...... ( -29.80) >DroSim_CAF1 48074 108 + 1 GCUGAUUAGCACAGCUAGUGGAUGGAUAGACA--UACAUAUAUAUA-AUAUAUCUGUCGCAUUAGCUUCAAUUUACUGGCGGU--------AACCUGGGCCGCGCCUUCCCGCAGAGAA ((.((...((..((((((((((((((((...(--((......))).-...)))))))).))))))))...........(((((--------.......)))))))..))..))...... ( -28.70) >DroEre_CAF1 45386 119 + 1 GCUGAUUAGCACAGCUAGUGGAUGGAUAGAUAUAUACAUAUAUGUAUAUGUAUCUGUCGCAUUAGCUUCAAUUUACUGGGGGGUGGGGGGCAACCUGUGCCGCGCCUUCCCGCAGAGAA (((....)))..((((((((((((((((.(((((((......))))))).)))))))).))))))))........(((.((((.(((.((((.....)))).).)).)))).))).... ( -49.40) >DroYak_CAF1 45682 107 + 1 ACUGAUUAGCACAGCUAGUGGGUAGAUGGAUAUAUACA----UGUAUAUAUAUCUGUCGCAUUAGCUUCAAUUUACUACGGGG--------AACGUGGGCCGCGCCUUCCCGCAGAGAA .(((........(((((((((..(((((.((((((...----.)))))).)))))..).))))))))...........(((((--------(.((((...))))..))))))))).... ( -33.90) >DroAna_CAF1 57966 80 + 1 ACUUAUUAGCAC-GUUAGUCGGU-------------------------UGUAUCUGUCGCAUUAACUUCAAUUUACGCUG-----------GAUCUG--CCCAGCACCCCCACUGACUA ............-((((((.((.-------------------------.(((..((..(......)..))...)))((((-----------(.....--.)))))...)).)))))).. ( -17.40) >consensus GCUGAUUAGCACAGCUAGUGGAUGGAUAGAUA__UACAUAUAUAUA_AUAUAUCUGUCGCAUUAGCUUCAAUUUACUGGCGGU________AACCUGGGCCGCGCCUUCCCGCAGAGAA .(((....((..((((((((((((((((......................)))))))).))))))))...........((((.................)))))).......))).... (-14.46 = -15.55 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:03 2006