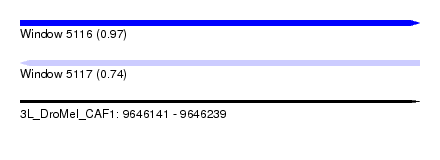
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,646,141 – 9,646,239 |
| Length | 98 |
| Max. P | 0.969249 |

| Location | 9,646,141 – 9,646,239 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.95 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9646141 98 + 23771897 UGUGGGGGUGUGGCA-AGAACACGCCCCAAAAAAAAAAACUCAAAUUGGUU--AGAGAA------ACGAGUUCCCUUUAUGGUCCCA---AUUCCGGACUUGGGGACUUG ....((((((((...-....))))))))...........(((.........--.)))..------.(((((((((.....(((((..---.....))))).))))))))) ( -36.00) >DroSec_CAF1 41703 102 + 1 UGUGGGGGUGUGGCA-AGAACACGCCCCGAA-----AAACUCAAAUUGGUU--AGAGAAACCAAAACGAGCUCCCUUUAUGGUCCCAAUAAUUCUGGACUUGGGGACUUG ....((((((((...-....))))))))...-----.........((((((--.....))))))..((((.(((((....(((((..........))))).))))))))) ( -37.00) >DroSim_CAF1 44249 102 + 1 UGUGGGGGUGUGGCA-AGAACACGCCCCGAA-----AAACUCAAAUUGGUU--AGAGAAACCAAAACGAGCUCCCUUUAUGGUCCCAAUAAUUCUGGACUUGGGGACUUG ....((((((((...-....))))))))...-----.........((((((--.....))))))..((((.(((((....(((((..........))))).))))))))) ( -37.00) >DroEre_CAF1 41515 95 + 1 UGUGGGGGUGUGGCA-AGAACACGCCCCGAA-----AAACUCAAAUUGGGA--AGAGAAACCAACACGAGCUCCCUUUAUGGGCCCA---ACUCUGGACUUGGGGA---- .(((((((((((...-....))))))))...-----.........((((..--.......))))))).....(((.....)))((((---(........)))))..---- ( -31.30) >DroYak_CAF1 41911 95 + 1 UGUGGGGGUGUGGCA-AGAACACGCCCCGAA-----AAACUCAAUUUGGGA--AGAGGAAACAAAACGAGCUCCCUUUAUGGGCUCC---AUUCUGGACUUGGGGA---- ....((((((((...-....))))))))...-----...(((((.((.(((--(..(....).....((((((.......)))))).---.)))).)).)))))..---- ( -35.30) >DroAna_CAF1 54092 102 + 1 UGUGGGGGUGUGGCAGAAAACAGGCCCAAAA--AUAAAGCUUAAAAUAGCACACAAGCAGCCUAAAAGAGCUCCCUUCAUGAGCCCAU--GUUGGGUACUUUGGAA---- ((((.((((.((........)).))))....--.....(((......))).))))...........((((..(((..((((....)))--)..)))..))))....---- ( -28.30) >consensus UGUGGGGGUGUGGCA_AGAACACGCCCCGAA_____AAACUCAAAUUGGUA__AGAGAAACCAAAACGAGCUCCCUUUAUGGGCCCA___AUUCUGGACUUGGGGA____ ....((((((((........))))))))...........(((............)))..............(((((....((.((..........)).)).))))).... (-19.36 = -19.95 + 0.59)



| Location | 9,646,141 – 9,646,239 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.92 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9646141 98 - 23771897 CAAGUCCCCAAGUCCGGAAU---UGGGACCAUAAAGGGAACUCGU------UUCUCU--AACCAAUUUGAGUUUUUUUUUUUGGGGCGUGUUCU-UGCCACACCCCCACA ...(..(((((........)---))))..)..((((((((((((.------......--........))))))))))))..(((((.((((...-....))))))))).. ( -27.06) >DroSec_CAF1 41703 102 - 1 CAAGUCCCCAAGUCCAGAAUUAUUGGGACCAUAAAGGGAGCUCGUUUUGGUUUCUCU--AACCAAUUUGAGUUU-----UUCGGGGCGUGUUCU-UGCCACACCCCCACA ...(..(((((...........)))))..).....(((((((((..((((((.....--))))))..)))))))-----)).((((.((((...-....))))))))... ( -33.30) >DroSim_CAF1 44249 102 - 1 CAAGUCCCCAAGUCCAGAAUUAUUGGGACCAUAAAGGGAGCUCGUUUUGGUUUCUCU--AACCAAUUUGAGUUU-----UUCGGGGCGUGUUCU-UGCCACACCCCCACA ...(..(((((...........)))))..).....(((((((((..((((((.....--))))))..)))))))-----)).((((.((((...-....))))))))... ( -33.30) >DroEre_CAF1 41515 95 - 1 ----UCCCCAAGUCCAGAGU---UGGGCCCAUAAAGGGAGCUCGUGUUGGUUUCUCU--UCCCAAUUUGAGUUU-----UUCGGGGCGUGUUCU-UGCCACACCCCCACA ----..(((((.(....).)---))))(((.....)))((((((.(((((.......--..))))).)))))).-----...((((.((((...-....))))))))... ( -31.80) >DroYak_CAF1 41911 95 - 1 ----UCCCCAAGUCCAGAAU---GGAGCCCAUAAAGGGAGCUCGUUUUGUUUCCUCU--UCCCAAAUUGAGUUU-----UUCGGGGCGUGUUCU-UGCCACACCCCCACA ----..........((((((---((..(((.....)))..).)))))))....(((.--.........)))...-----...((((.((((...-....))))))))... ( -24.90) >DroAna_CAF1 54092 102 - 1 ----UUCCAAAGUACCCAAC--AUGGGCUCAUGAAGGGAGCUCUUUUAGGCUGCUUGUGUGCUAUUUUAAGCUUUAU--UUUUGGGCCUGUUUUCUGCCACACCCCCACA ----.....(((((.((...--..((((((.......)))))).....)).)))))(((((((......))).....--.....(((..(....).)))))))....... ( -22.40) >consensus ____UCCCCAAGUCCAGAAU___UGGGACCAUAAAGGGAGCUCGUUUUGGUUUCUCU__AACCAAUUUGAGUUU_____UUCGGGGCGUGUUCU_UGCCACACCCCCACA .....(((................))).((.....))(((((((..((((...........))))..)))))))........((((.((((........))))))))... (-16.59 = -17.92 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:01 2006