
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,643,496 – 9,643,611 |
| Length | 115 |
| Max. P | 0.605611 |

| Location | 9,643,496 – 9,643,611 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
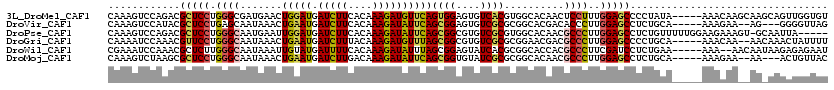
>3L_DroMel_CAF1 9643496 115 + 23771897 CAAAGUCCAGACGCUCCUGGGCGAUGAACUGGAUGAUCUUCACAAAGAUGUUCAGUGGAGUGUCACGUGGCACAACUCCUUUGGAGCCCCUAUA-----AAACAAGCAAGCAGUUGGUGU ....((((((......)))))).((.(((((..(((((((....)))))(((..((((.(((((....)))))..((((...))))...)))).-----.)))...))..))))).)).. ( -31.10) >DroVir_CAF1 41249 110 + 1 CAAAGUCCAUACGCUCCUGAGCAAUAAACUGAAUGAUCUUCACAAAGAUAUUCAGCGGAGUGUCGCGCGGCACGACACCCUUGGAGCCUCUGCA-----AAAGAA--AG---GGGGUUAG ..((.(((....(((((...........(((((((.((((....))))))))))).((.((((((.......))))))))..))))).(((...-----..))).--..---))).)).. ( -32.20) >DroPse_CAF1 41125 114 + 1 CAAAGUCCAGACGCUCCUGGGCAAUGAAUUGGAUGAUCUUCACAAAGAUAUUCAGCGGCGUGUCGCGUGGCACAACGCCCUUGGAGCCUCUGUUUUUGGAAGAAAGU-GCAAUUA----- (((((..((((.(((((.((((.......((((((.((((....))))))))))((.(((.....))).)).....))))..))))).)))).))))).........-.......----- ( -39.10) >DroGri_CAF1 37177 113 + 1 CAAAAUCCAAACGUUCCUGGGCAAUAAACUGAAUGAUCUUUACAAAGAUGUUUAGCGGCGUGUCGCGCGGAACGACGCCCUUGGAGCCCCUGCA-----AAACAA--AACAAACUAUUUU ............(((((.((((......((((((.(((((....)))))))))))(.(((.....))).)......))))..))))).......-----......--............. ( -27.20) >DroWil_CAF1 59285 113 + 1 CGAAAUCCAAACGCUCUUGGGCAAUAAAUUGUAUGAUUUUCACAAAGAUAUUUAGCGGAGUAUCACGCGGCACCACGCCCUUCGAUCCUCUGAA-----AAA--AACAAUAAGAGAGAAU ((((..((((......))))((......((((.........)))).(((((((....)))))))..))(((.....))).)))).((((((...-----...--.......)))).)).. ( -19.62) >DroMoj_CAF1 41227 110 + 1 CAAAGUCUAAGCGCUCCUGGGCAAUAAACUGAAUGAUCUUGACAAAGAUAUUCAGCGGUGUAUCGCGCGGCACAACGCCCUUGGAGCCUCUGCA-----AAAGAA--AA---ACUGUUAC .........((((((((.((((.......((((((.((((....))))))))))((.(((.....))).)).....))))..))))).(((...-----..))).--..---...))).. ( -33.80) >consensus CAAAGUCCAAACGCUCCUGGGCAAUAAACUGAAUGAUCUUCACAAAGAUAUUCAGCGGAGUGUCGCGCGGCACAACGCCCUUGGAGCCUCUGCA_____AAACAA__AA_AAGCUGUUAU ............(((((.((((.......(((((.(((((....))))))))))((((....))))..........))))..)))))................................. (-19.62 = -19.57 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:00 2006