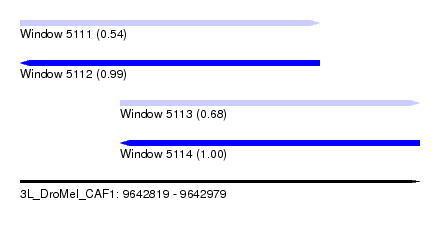
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,642,819 – 9,642,979 |
| Length | 160 |
| Max. P | 0.996394 |

| Location | 9,642,819 – 9,642,939 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9642819 120 + 23771897 GUUGAGGAAAGUGAACAGGAUGCGCGUGCAGUUCUCCAGCAGCUGCGGAUAGGUGUCGGUGACGUCGCGCACCAGAAACUGAGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGGAA ((((..(((.(....)....(((....))).)))..)))).....(((((.((((((((.((.((((.((((.....((.....))....)))).)))).)))))))))).))))).... ( -44.80) >DroVir_CAF1 40578 120 + 1 GUUUAGAAAGACAAACAAGAUGCGCGUGCAGUUCUCCAACAAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAA (((.((((............(((....))).))))..)))..((.(((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))).)). ( -35.92) >DroGri_CAF1 36506 120 + 1 GUUUAGGAAGACAAACAAGAUGCGCGUGCAGUUCUCCAACAAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAA .....(((..((........(((....)))))..))).....((.(((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))).)). ( -39.20) >DroWil_CAF1 58614 120 + 1 AUUUAAAAAUACAAACAAUAUACGCGUACAAUUCUCCAAUAAUUGCGGAUAGGUGUCGGUGACAUCGCGCACUAGAAACUGUGUGUAACCGUGCACAACAUCCUGACGCCAAUCCGGAAA ..........................................((.(((((.((((((((.((....((((((........))))))....((.....)).)))))))))).))))).)). ( -31.10) >DroMoj_CAF1 40559 120 + 1 GGUUAGGAGAACAAACAAGAUGCGCGUACAGUUCUCCAACAAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAA .(((.(((((((..................))))))))))..((.(((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))).)). ( -44.17) >DroAna_CAF1 51045 120 + 1 GUUGAGAAAGUUGAACAAGAUACGCGUGCAGUUCUCCAGGAGUUGCGGAUAGGUGUCGGUGACGUCGCGCACCAGAAACUGGGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAA ...(((((...((.((.........)).)).)))))......((.(((((.((((((((.((.((((.(((((((...)))((.....)))))).)))).)))))))))).))))).)). ( -43.50) >consensus GUUUAGAAAGACAAACAAGAUGCGCGUGCAGUUCUCCAACAAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAA ...........................((((((.......))))))((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))..... (-32.28 = -32.28 + 0.00)


| Location | 9,642,819 – 9,642,939 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -37.65 |
| Energy contribution | -36.10 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9642819 120 - 23771897 UUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCUGCUGGAGAACUGCACGCGCAUCCUGUUCACUUUCCUCAAC .(((.((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((....)).)))(((((((....)))....))))............ ( -44.60) >DroVir_CAF1 40578 120 - 1 UUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGCACGCGCAUCUUGUUUGUCUUUCUAAAC ....(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))......(((((((...(((.(((.....))).))).))))))).. ( -41.10) >DroGri_CAF1 36506 120 - 1 UUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGCACGCGCAUCUUGUUUGUCUUCCUAAAC ....(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))......((((.(((..(((.(((.....))).))).))))))).. ( -40.00) >DroWil_CAF1 58614 120 - 1 UUUCCGGAUUGGCGUCAGGAUGUUGUGCACGGUUACACACAGUUUCUAGUGCGCGAUGUCACCGACACCUAUCCGCAAUUAUUGGAGAAUUGUACGCGUAUAUUGUUUGUAUUUUUAAAU .....((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))((((((.......))))))....(((((.....)))))........ ( -35.00) >DroMoj_CAF1 40559 120 - 1 UUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGUACGCGCAUCUUGUUUGUUCUCCUAACC ....(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).))))).....((((((((((...(((.......)))..))))))).))). ( -46.80) >DroAna_CAF1 51045 120 - 1 UUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAACUCCUGGAGAACUGCACGCGUAUCUUGUUCAACUUUCUCAAC ....(((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).))))).........(((((......(((.....)))......)))))... ( -42.70) >consensus UUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUUGUUGGAGAACUGCACGCGCAUCUUGUUUGUCUUCCUAAAC ....(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))........((((((((((....)))....))))....)))..... (-37.65 = -36.10 + -1.55)


| Location | 9,642,859 – 9,642,979 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -45.37 |
| Consensus MFE | -36.48 |
| Energy contribution | -36.23 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9642859 120 + 23771897 AGCUGCGGAUAGGUGUCGGUGACGUCGCGCACCAGAAACUGAGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGGAAAUCUAUGACCAAAGUCGUAAGCGACUGGUGGGUUAGAAUU .....(((((.((((((((.((.((((.((((.....((.....))....)))).)))).)))))))))).))))).....((((...(((.(((((....)))))..)))..))))... ( -50.20) >DroVir_CAF1 40618 120 + 1 AAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAAGUCUAUGACCAGCGUCUGCAGCGAUUGAUGCGUCAAUAUG ..((.(((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))).))......((((..(((((..........)))))))))..... ( -42.00) >DroWil_CAF1 58654 120 + 1 AAUUGCGGAUAGGUGUCGGUGACAUCGCGCACUAGAAACUGUGUGUAACCGUGCACAACAUCCUGACGCCAAUCCGGAAAAUCAAUGACCAGCGUCUGCAGCGAUUGAUGCGUCAAUAUG ..((.(((((.((((((((.((....((((((........))))))....((.....)).)))))))))).))))).))......((((..(((((..........)))))))))..... ( -39.70) >DroMoj_CAF1 40599 120 + 1 AAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAAGUCUUUGACCAGCGUCUGCAGCGAUUGAUACGUCAGUAUG ((((((((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))...........(((....)))....)))))).((((....)))). ( -39.00) >DroAna_CAF1 51085 120 + 1 AGUUGCGGAUAGGUGUCGGUGACGUCGCGCACCAGAAACUGGGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAAGUCAAUCACCAGCGUGGUCAGCGAUUGGUGCGUCAAAAUG ..((.(((((.((((((((.((.((((.(((((((...)))((.....)))))).)))).)))))))))).))))).)).......(((((((((.....))).)))))).......... ( -49.20) >DroPer_CAF1 39635 120 + 1 AGCUGCGGAUAGGUGUCGGUGACGUCGCGCACCAGAAACUGGGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGGAAGUCAAUGACCAGCGUCUGCAGCGACUGGUGCGUCAGUAUG .(((((((((.((((((((.((.((((.(((((((...)))((.....)))))).)))).)))))))))).....((..(.....)..))...))))))))).(((((....)))))... ( -52.10) >consensus AAUUGCGGAUAGGUGUCGGUGACAUCGCGCACCAGAAACUGUGUGUAACCGUGCACGACAUCCUGACGCCAAUCCGGAAAGUCAAUGACCAGCGUCUGCAGCGAUUGAUGCGUCAAUAUG ((((((((((.((((((((.((..(((.((((.....((.....))....)))).)))..)))))))))).))))...........(((....)))....)))))).............. (-36.48 = -36.23 + -0.25)


| Location | 9,642,859 – 9,642,979 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -40.18 |
| Energy contribution | -39.52 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9642859 120 - 23771897 AAUUCUAACCCACCAGUCGCUUACGACUUUGGUCAUAGAUUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACUCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCU ...((((..(((..(((((....))))).)))...)))).....(((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))..... ( -46.90) >DroVir_CAF1 40618 120 - 1 CAUAUUGACGCAUCAAUCGCUGCAGACGCUGGUCAUAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUU ...(((((....)))))...(((.(((....)))...........((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))))... ( -40.50) >DroWil_CAF1 58654 120 - 1 CAUAUUGACGCAUCAAUCGCUGCAGACGCUGGUCAUUGAUUUUCCGGAUUGGCGUCAGGAUGUUGUGCACGGUUACACACAGUUUCUAGUGCGCGAUGUCACCGACACCUAUCCGCAAUU .........((((((((.(((((....)).))).)))))).....((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))).... ( -41.90) >DroMoj_CAF1 40599 120 - 1 CAUACUGACGUAUCAAUCGCUGCAGACGCUGGUCAAAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGAUGUCACCGACACCUAUCCGCAAUU .((((....)))).......(((.(((....)))...........((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))))... ( -40.00) >DroAna_CAF1 51085 120 - 1 CAUUUUGACGCACCAAUCGCUGACCACGCUGGUGAUUGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAACU .........((..((((((((.........)))))))).......((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).)))))).... ( -47.80) >DroPer_CAF1 39635 120 - 1 CAUACUGACGCACCAGUCGCUGCAGACGCUGGUCAUUGACUUCCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACCCAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAGCU ......(((......)))(((((.(((....)))...........((((.((.(((.(((((((((((((((.....))(((...))))))))))))))).).))).)).))))))))). ( -52.30) >consensus CAUACUGACGCACCAAUCGCUGCAGACGCUGGUCAUAGACUUUCCGGAUUGGCGUCAGGAUGUCGUGCACGGUUACACACAGUUUCUGGUGCGCGACGUCACCGACACCUAUCCGCAACU ..................((....(((....)))...........((((.((.(((.(((((((((((((((..((.....))..)).)))))))))))).).))).)).)))))).... (-40.18 = -39.52 + -0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:59 2006