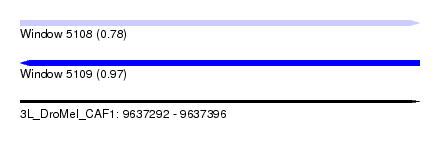
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,637,292 – 9,637,396 |
| Length | 104 |
| Max. P | 0.965013 |

| Location | 9,637,292 – 9,637,396 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9637292 104 + 23771897 AU---------------AUAUAUUUAACUUGCCUGCAGGGCGCUGAACUGCAGCUUCUCCUCCUCGAGCGUAUGGUCGCUCUGCAUGGUGUAACCAAGUGGCUUGGAGAAGCUGCCGCA ..---------------..........((((....))))(((.......(((((((((((.((..(((((......))))).((.(((.....))).))))...)))))))))))))). ( -39.81) >DroVir_CAF1 34610 107 + 1 -------C-----AGCUCCAGCUGCGACUUGCCUGUAAAGCGCUAAAUUGCAGCUUCUCCUCCUCGAGAGUUUGGCCAAUCUGUGAGCUAAUGCUUAACGGCUUAGAGAAGCUGCCGCA -------.-----(((....)))(((....((((....)).))......(((((((((((((.....)))...((((......(((((....)))))..))))..))))))))))))). ( -36.20) >DroPse_CAF1 33654 112 + 1 -------UGGGUUCGUUGUGGAUUAUACUUGCCUGCAGGGCGCUGAACUGCAGCUUCUCCUCCUCGAGGGUCUGGCCAAUCUGCGAUGUAAUGCUCAGCGGCUUGGAGAAGCUGCCGCA -------(((((.(((((..((((....(((....)))(((((((.....))))...(((((...)))))....)))))))..)))))....)))))(((((...........))))). ( -39.90) >DroYak_CAF1 33245 119 + 1 AAAACGUUGCGUUUUUUUGGUAUUUAACUUGCCUGCAGGGCGCUGAACUGCAGCUUCUCCUCCUCGAGCGUAUGGUCGCUCUGCAUGGUGUACCCAAGUGGCUUGGAGAAGCUGCCGCA .....((((((((((...((((.......))))...)))))))..))).(((((((((((.((..(((((......))))).((.(((.....))).))))...))))))))))).... ( -44.60) >DroAna_CAF1 45138 107 + 1 ----------GAC-UUUCGGCA-UUAACUUGCCUGCAAGGCACUAAACUGCAGCUUCUCCUCCUCAAGGGUCUGGCCAAUCUGCGAGGUAAUGCUCAGCGGCUUUGAGAAGCUGCCGCA ----------...-....((((-......)))).((.((........))((((((((((.(((....)))...((((.....(((((......))).))))))..)))))))))).)). ( -34.00) >DroPer_CAF1 33699 112 + 1 -------UGGGUUCGUUGUGGAUUAUACUUGCCUGCAGGGCGCUGAACUGCAGCUUCUCCUCCUCGAGGGUCUGGCCAAUCUGCGAUGUAAUGCUCAGCGGCUUGGAGAAGCUGCCGCA -------(((((.(((((..((((....(((....)))(((((((.....))))...(((((...)))))....)))))))..)))))....)))))(((((...........))))). ( -39.90) >consensus _______U__GUU_GUUCUGGAUUUAACUUGCCUGCAGGGCGCUGAACUGCAGCUUCUCCUCCUCGAGGGUCUGGCCAAUCUGCGAGGUAAUGCUCAGCGGCUUGGAGAAGCUGCCGCA ...........................((((....))))(((.......(((((((((((.............((((.....((........)).....)))).)))))))))))))). (-25.25 = -25.58 + 0.34)

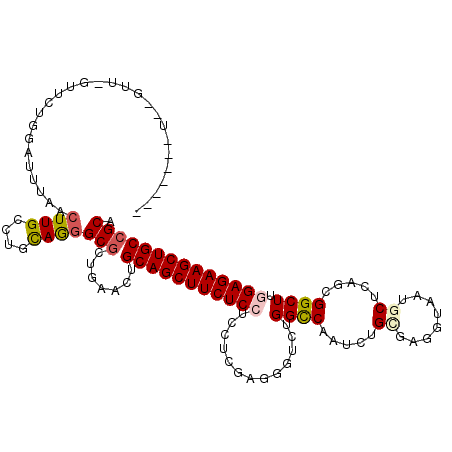
| Location | 9,637,292 – 9,637,396 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.98 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9637292 104 - 23771897 UGCGGCAGCUUCUCCAAGCCACUUGGUUACACCAUGCAGAGCGACCAUACGCUCGAGGAGGAGAAGCUGCAGUUCAGCGCCCUGCAGGCAAGUUAAAUAUAU---------------AU .((.(((((((((((...((...(((.....)))....(((((......)))))..)).)))))))))))......))(((.....))).............---------------.. ( -39.80) >DroVir_CAF1 34610 107 - 1 UGCGGCAGCUUCUCUAAGCCGUUAAGCAUUAGCUCACAGAUUGGCCAAACUCUCGAGGAGGAGAAGCUGCAAUUUAGCGCUUUACAGGCAAGUCGCAGCUGGAGCU-----G------- .(((((((((((((...((((...(((....))).......))))....(((.....)))))))))))))........(((.....)))....)))(((....)))-----.------- ( -35.90) >DroPse_CAF1 33654 112 - 1 UGCGGCAGCUUCUCCAAGCCGCUGAGCAUUACAUCGCAGAUUGGCCAGACCCUCGAGGAGGAGAAGCUGCAGUUCAGCGCCCUGCAGGCAAGUAUAAUCCACAACGAACCCA------- ....(((((((((((..((((....((........))....)))).....((....)).))))))))))).((((...(((.....)))..((.......))...))))...------- ( -35.90) >DroYak_CAF1 33245 119 - 1 UGCGGCAGCUUCUCCAAGCCACUUGGGUACACCAUGCAGAGCGACCAUACGCUCGAGGAGGAGAAGCUGCAGUUCAGCGCCCUGCAGGCAAGUUAAAUACCAAAAAAACGCAACGUUUU (((((((((((((((...((...(((.....)))....(((((......)))))..)).)))))))))))........(((.....)))...................))))....... ( -44.80) >DroAna_CAF1 45138 107 - 1 UGCGGCAGCUUCUCAAAGCCGCUGAGCAUUACCUCGCAGAUUGGCCAGACCCUUGAGGAGGAGAAGCUGCAGUUUAGUGCCUUGCAGGCAAGUUAA-UGCCGAAA-GUC---------- .(((((((((((((...((......))....(((((.((...((.....)))))))))..)))))))))).......((((.....))))......-)))(....-)..---------- ( -34.10) >DroPer_CAF1 33699 112 - 1 UGCGGCAGCUUCUCCAAGCCGCUGAGCAUUACAUCGCAGAUUGGCCAGACCCUCGAGGAGGAGAAGCUGCAGUUCAGCGCCCUGCAGGCAAGUAUAAUCCACAACGAACCCA------- ....(((((((((((..((((....((........))....)))).....((....)).))))))))))).((((...(((.....)))..((.......))...))))...------- ( -35.90) >consensus UGCGGCAGCUUCUCCAAGCCGCUGAGCAUUACCUCGCAGAUUGGCCAGACCCUCGAGGAGGAGAAGCUGCAGUUCAGCGCCCUGCAGGCAAGUUAAAUCCACAAC_AAC__A_______ .((.(((((((((((..((......))....(((((.((............))))))).)))))))))))......))(((.....))).............................. (-27.48 = -27.98 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:54 2006