
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,636,462 – 9,636,597 |
| Length | 135 |
| Max. P | 0.983696 |

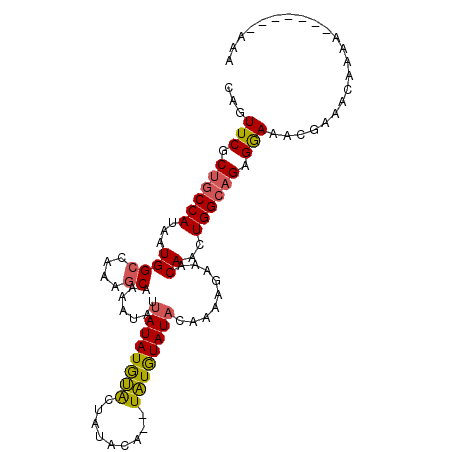
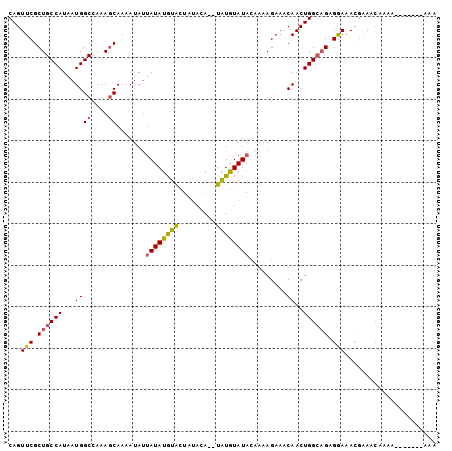
| Location | 9,636,462 – 9,636,565 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9636462 103 + 23771897 CAGUUCGCUGCCAUAAUGGCCAAAGCAAAAUAUUAUAUGCACUUUAUAUAUGUGUAUACAAAAGAAACAACUGGCAGAGGAAAUGAAACAAAAAAAAAAAAAA ((.(((.((((((...((((....)).........(((((((.........))))))).........))..))))))..))).)).................. ( -19.20) >DroSec_CAF1 32265 94 + 1 CAGUUCGCUGCCAUAAUGGCCAAAGCAAAAUAUUAUAUGUACUAUACA--UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACACAA-------AAA ...(((.((((((...((((....))...((((.(((((((...))))--)))))))..........))..)))))).(....))))......-------... ( -17.70) >DroSim_CAF1 34791 94 + 1 CAGUUCGCUGCCAUAAUGGCCAAAGCAAAAUAUUAUAUGUACUAUAUA--UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACACAA-------AAA ...(((.((((((...((((....)).......((((((((.....))--))))))...........))..)))))).(....))))......-------... ( -16.60) >DroYak_CAF1 32441 88 + 1 CAGUUCGCUGCCAUAAUGGCCAAACCAAAAUAAUAUAUAUGC---ACA--UAUAUAU---AAAGAAACAACUGGACGAGAAAAUACAAAAAGA-------AAA (((((..(((((.....)))............((((((((..---..)--)))))))---..))....)))))....................-------... ( -12.10) >consensus CAGUUCGCUGCCAUAAUGGCCAAAGCAAAAUAUUAUAUGUACUAUACA__UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACAAAA_______AAA ...(((.((((((...((((....)).......((((((((.........)))))))).........))..)))))).)))...................... (-11.23 = -11.10 + -0.13)

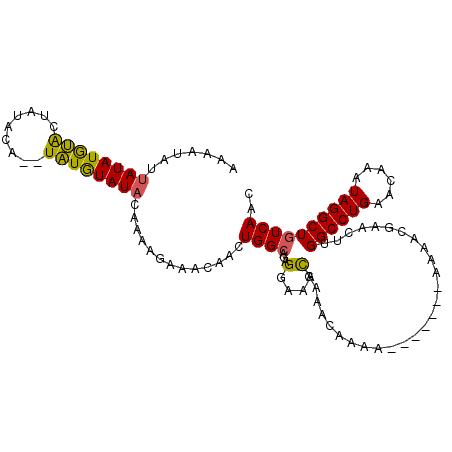
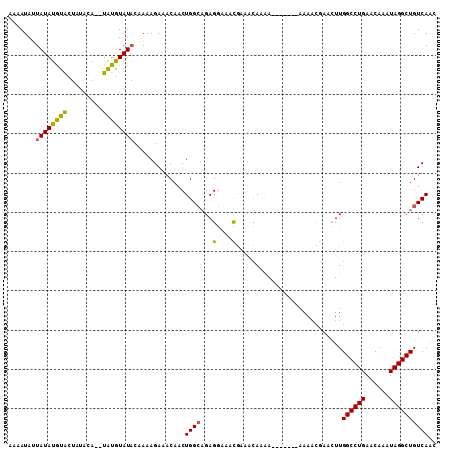
| Location | 9,636,488 – 9,636,597 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -14.78 |
| Consensus MFE | -9.76 |
| Energy contribution | -9.07 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9636488 109 + 23771897 AAAAUAUUAUAUGCACUUUAUAUAUGUGUAUACAAAAGAAACAACUGGCAGAGGAAAUGAAACAAAAAAAAAAAAAAACGAACUUGGCCUGAACAAAUAGGCUGUCAAC .........(((((((.........))))))).............((((.(((.............................)))((((((......)))))))))).. ( -15.05) >DroSec_CAF1 32291 100 + 1 AAAAUAUUAUAUGUACUAUACA--UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACACAA-------AAAACGAACUUGGCCUGAACAAAUAGGCUGUCAAC ...((((.(((((((...))))--)))))))..............((((...(....).........-------...........((((((......)))))))))).. ( -16.40) >DroSim_CAF1 34817 100 + 1 AAAAUAUUAUAUGUACUAUAUA--UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACACAA-------AAAACGAACUUGGCCUGAACAAAUAGGCUGUCAAC .......((((((((.....))--))))))...............((((...(....).........-------...........((((((......)))))))))).. ( -15.30) >DroYak_CAF1 32467 92 + 1 AAAAUAAUAUAUAUGC---ACA--UAUAUAU---AAAGAAACAACUGGACGAGAAAAUACAAAAAGA-------AAA--AAGGUUGGCCUGAACAAAUAGGCUGUCAGC ......((((((((..---..)--)))))))---...((..(((((.....................-------...--..)))))(((((......)))))..))... ( -12.37) >consensus AAAAUAUUAUAUGUACUAUACA__UAUGUAUACAAAAGAAACAACUGGCAGAGGAAACGAAACAAAA_______AAAACGAACUUGGCCUGAACAAAUAGGCUGUCAAC .......((((((((.........)))))))).............((((...(....)...........................((((((......)))))))))).. ( -9.76 = -9.07 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:52 2006