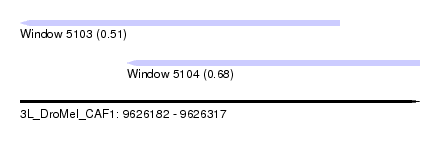
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,626,182 – 9,626,317 |
| Length | 135 |
| Max. P | 0.684818 |

| Location | 9,626,182 – 9,626,290 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
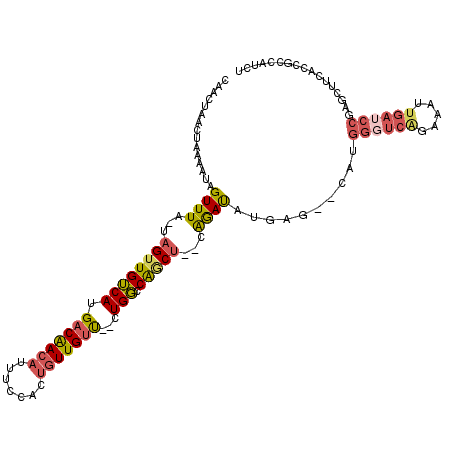
>3L_DroMel_CAF1 9626182 108 - 23771897 CAACUAACUAAAAUAGUUUA-UAGUUGUCAUGACAACAUUUCCACUGUUGUU--CUGGCCGACU--UAGAUAUGAG--CAUGGGUCAGAAAUUGAUCCGAGCUCCACCGCCAUCU ...............(((((-.((((((((.(((((((.......)))))))--.))).)))))--)))))..(((--(.((((((((...)))))))).))))........... ( -29.10) >DroSec_CAF1 21974 108 - 1 CAACUAAGUAAAAUAGUUUA-UAGUUGUCAUGACAACAAUUCGACUGUUGUU--CUGGCCGUCU--CAGAUAUGAG--CAGGGGUCGGAAAUUGAUCCGAGCUUCACCGCCAUCU ......((...(((((((.(-(.(((((....))))).))..)))))))...--))(((.(.((--((....))))--).(((((((((......))))).))))...))).... ( -28.80) >DroSim_CAF1 24466 108 - 1 CAACUAACUAAAAUAGUUUA-UAGUUGUCAUGACAACAUUUCGACUGUUGUU--CUGGCCGACU--CAAAUAUGAG--CAUGGGUCGGAAAUUGAUCCGAGCUUCACCGCCAUCU ((((((((((...))))...-))))))....(((((((.......)))))))--.((((.(.((--((....))))--)..((.(((((......))))).)).....))))... ( -26.10) >DroEre_CAF1 21263 108 - 1 CAAGCAGCUAAAAUAGUUUA-UAGUUGUCAUGCCAACAUUUCCACUGUUGUU--CUGGCCAGCU--CAGAUAUGAG--CAUGGGUCAGAAAUUGAUCCCAGCUUGAACGCCAUCU ((((((((((...)))))..-..(..((((...(((((.......)))))((--((((((.(((--((....))))--)...))))))))..))))..).))))).......... ( -31.80) >DroYak_CAF1 22018 110 - 1 CAAGCAACUAAAAUAGUUUA-UAGUUGUCAUGACGACAUUUCCACUGUUGUC--CUGGCCAGCU--CAGAUAUGAGGGUAUGGUUCAGAAAUUGAUCCGAGCCUCAACGCCAUCU ...(((((((.((....)).-)))))))...(((((((.......)))))))--.((((...((--((....))))(((.(((.((((...)))).))).))).....))))... ( -29.50) >DroAna_CAF1 34376 93 - 1 ------CUUAAAAUAGUUUGGCCGUUGGCAUGACAACUUUAUGACUCUUGUUUGUUGUCCAGCUUUUGGACGUAAG--CC-GGGCCAGC-------------AUCCCACUCAUCC ------.........(..(((..((((((..((((((.....(((....))).))))))......((((.(....)--))-))))))))-------------...)))..).... ( -24.30) >consensus CAACUAACUAAAAUAGUUUA_UAGUUGUCAUGACAACAUUUCCACUGUUGUU__CUGGCCAGCU__CAGAUAUGAG__CAUGGGUCAGAAAUUGAUCCGAGCUUCACCGCCAUCU ...............((((...((((((((.(((((((.......)))))))...))).)))))...))))..........((((((.....))))))................. (-13.77 = -14.22 + 0.45)

| Location | 9,626,218 – 9,626,317 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -13.10 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
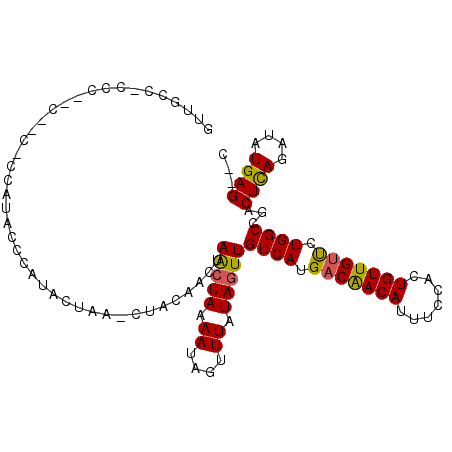
>3L_DroMel_CAF1 9626218 99 - 23771897 AUUGGC---------CACCAUACCCAUACUAAGCUACAACUAACUAAAAUAGUUUAUAGUUGUCAUGACAACAUUUCCACUGUUGUUCUGGCCGACUUAGAUAUGAG--C .(((((---------((..................(((((((((((...))))...)))))))...(((((((.......))))))).)))))))((((....))))--. ( -22.20) >DroSec_CAF1 22010 107 - 1 GUUGCC-CCCACCCCCGCCAUACUCAUACUAAACUACAACUAAGUAAAAUAGUUUAUAGUUGUCAUGACAACAAUUCGACUGUUGUUCUGGCCGUCUCAGAUAUGAG--C ......-...............((((((((.(((((.(((((.......)))))..)))))((((.(((((((.......))))))).))))......)).))))))--. ( -20.80) >DroSim_CAF1 24502 108 - 1 GUUGUCACCCACCCCCCCCAUACUCAUACUAAACUACAACUAACUAAAAUAGUUUAUAGUUGUCAUGACAACAUUUCGACUGUUGUUCUGGCCGACUCAAAUAUGAG--C ......................((((((.(((((((.............))))))).((((((((.(((((((.......))))))).))).)))))....))))))--. ( -20.42) >DroEre_CAF1 21299 88 - 1 GUUG----------------GCCCCAUCCU----CUCAAGCAGCUAAAAUAGUUUAUAGUUGUCAUGCCAACAUUUCCACUGUUGUUCUGGCCAGCUCAGAUAUGAG--C ..((----------------(((.......----.....(((((((.((....)).))))))).....(((((.......)))))....)))))(((((....))))--) ( -22.80) >DroYak_CAF1 22054 103 - 1 GCAGCU-CCCGUC--CUCGAUCCCCAUCCU----CCCAAGCAACUAAAAUAGUUUAUAGUUGUCAUGACGACAUUUCCACUGUUGUCCUGGCCAGCUCAGAUAUGAGGGU ......-....((--(((((((........----.......(((((...)))))...((((((((.(((((((.......))))))).))).)))))..))).)))))). ( -22.90) >consensus GUUGCC_CCC__C__C_CCAUACCCAUACUAA_CUACAACUAACUAAAAUAGUUUAUAGUUGUCAUGACAACAUUUCCACUGUUGUUCUGGCCGACUCAGAUAUGAG__C .........................................(((((.((....)).)))))((((.(((((((.......))))))).))))...((((....))))... (-13.10 = -12.86 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:49 2006