| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,624,130 – 9,624,224 |
| Length | 94 |
| Max. P | 0.844337 |

| Location | 9,624,130 – 9,624,224 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.61 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
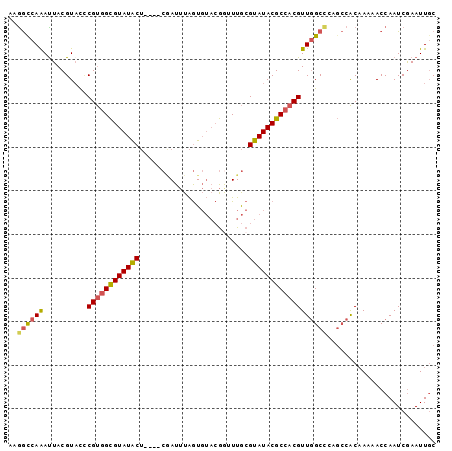
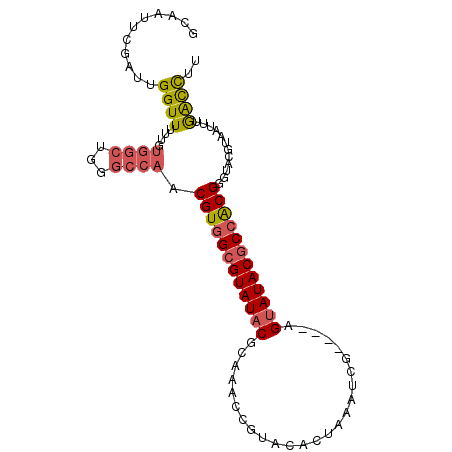
>3L_DroMel_CAF1 9624130 94 + 23771897 AAGGUCAAAUUACGUACCCGUGGCGUAUACU----CGAUUUAGUGUACGGUUUGCGUAUACGCCACGUUGGCCCAGCCACAAAAACCAAUCGAAUUGC ..(((.............((((((((((((.----(((....(....)...))).)))))))))))).((((...)))).....)))........... ( -28.20) >DroSec_CAF1 19811 94 + 1 AAGGUCAAAUUACGUACCCGUGGCGUAUACU----CGAUUUAGUGUACGGUUUGCGUAUACGCCACGUUGGCUAAGCCACAAAAACCAAUCAAAUUGC ..(((.............((((((((((((.----(((....(....)...))).)))))))))))).((((...)))).....)))........... ( -28.20) >DroSim_CAF1 22259 94 + 1 AAGGUCAAAUUACGUUCCCGUGGCGUAUACU----CGAUUUAGUGUACGAGUUGCGUAUACGCCACGUUGGCUAAGCCACAAAAACCAAUCGAAUUGC ..(((.............((((((((((((.----((((((.......)))))).)))))))))))).((((...)))).....)))........... ( -31.20) >DroEre_CAF1 19242 94 + 1 AAGGCCAAAUUACGUACUCGUGGCGUAUACU----CAAUUUAGUGUACGGCUUGCGUAUACGCCACGUUGGCCCAGCCACAAAAACCAAUCGAAUUGC ..((((((..........((((((((((((.----(((....(....)...))).))))))))))))))))))......................... ( -29.50) >DroYak_CAF1 20000 94 + 1 AAGGCCAAACUACGUACUCGUGGCGUAUACU----CAAUUUAGUGUACGGUUUGCGUAUACGCCACGUUGGCCCAGCCACAAAAACCAAUCGAAUUGC ..((((((..........((((((((((((.----(((....(....)...))).))))))))))))))))))......................... ( -30.20) >DroAna_CAF1 32575 88 + 1 CAAACUAGAUUACGUAUACGCCGUGUAUGCAAGCACCAUCUGAU----AAAUUACGUAUACGCCACGUUCGCC-----GC-AAAACCAAUCGAAUUUC .......((((..(((((((..((((......))))(....)..----......)))))))((..(....)..-----))-......))))....... ( -12.10) >consensus AAGGCCAAAUUACGUACCCGUGGCGUAUACU____CGAUUUAGUGUACGGUUUGCGUAUACGCCACGUUGGCCCAGCCACAAAAACCAAUCGAAUUGC ..((((((..........((((((((((((.........................))))))))))))))))))......................... (-20.61 = -20.61 + 0.00)


| Location | 9,624,130 – 9,624,224 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
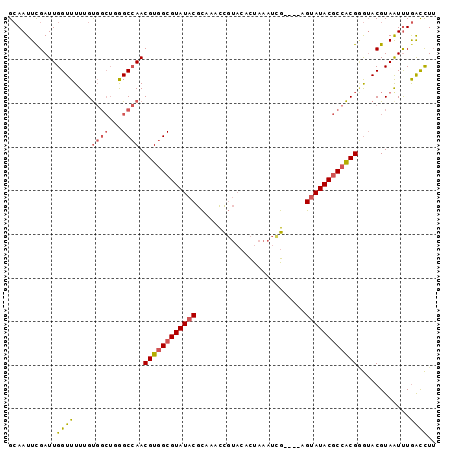
>3L_DroMel_CAF1 9624130 94 - 23771897 GCAAUUCGAUUGGUUUUUGUGGCUGGGCCAACGUGGCGUAUACGCAAACCGUACACUAAAUCG----AGUAUACGCCACGGGUACGUAAUUUGACCUU .........(((((((........)))))))((((((((((((....................----.))))))))))))(((.((.....))))).. ( -28.10) >DroSec_CAF1 19811 94 - 1 GCAAUUUGAUUGGUUUUUGUGGCUUAGCCAACGUGGCGUAUACGCAAACCGUACACUAAAUCG----AGUAUACGCCACGGGUACGUAAUUUGACCUU ...........((((....((((...)))).((((((((((((....................----.))))))))))))............)))).. ( -27.40) >DroSim_CAF1 22259 94 - 1 GCAAUUCGAUUGGUUUUUGUGGCUUAGCCAACGUGGCGUAUACGCAACUCGUACACUAAAUCG----AGUAUACGCCACGGGAACGUAAUUUGACCUU ...........((((....((((...)))).((((((((((.....(((((..........))----)))))))))))))............)))).. ( -28.10) >DroEre_CAF1 19242 94 - 1 GCAAUUCGAUUGGUUUUUGUGGCUGGGCCAACGUGGCGUAUACGCAAGCCGUACACUAAAUUG----AGUAUACGCCACGAGUACGUAAUUUGGCCUU ((....(((.......)))..)).(((((((((((((((((((.(((.............)))----.))))))))))))..........))))))). ( -32.12) >DroYak_CAF1 20000 94 - 1 GCAAUUCGAUUGGUUUUUGUGGCUGGGCCAACGUGGCGUAUACGCAAACCGUACACUAAAUUG----AGUAUACGCCACGAGUACGUAGUUUGGCCUU ((....(((.......)))..)).(((((((((((((((((((.(((.............)))----.))))))))))))..........))))))). ( -31.32) >DroAna_CAF1 32575 88 - 1 GAAAUUCGAUUGGUUUU-GC-----GGCGAACGUGGCGUAUACGUAAUUU----AUCAGAUGGUGCUUGCAUACACGGCGUAUACGUAAUCUAGUUUG .(((((.(((((.....-((-----.((....)).))((((((((....(----(((((.......))).)))....)))))))).))))).))))). ( -20.30) >consensus GCAAUUCGAUUGGUUUUUGUGGCUGGGCCAACGUGGCGUAUACGCAAACCGUACACUAAAUCG____AGUAUACGCCACGGGUACGUAAUUUGACCUU ...........((((....((((...)))).((((((((((((.........................))))))))))))............)))).. (-20.80 = -21.16 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:47 2006