
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,618,907 – 9,619,024 |
| Length | 117 |
| Max. P | 0.815807 |

| Location | 9,618,907 – 9,619,024 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -41.14 |
| Consensus MFE | -29.17 |
| Energy contribution | -30.31 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
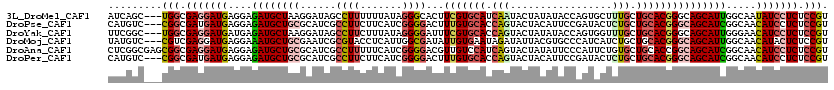
>3L_DroMel_CAF1 9618907 117 + 23771897 AUCAGC---UGGCGAGGAUGAGGAGAUGCUAAGGAUAGCCUUUUUUAUAGGGCACUUCGUGCAUCAAUACUAUAUACCAGUGCUUUGCUGCACGGGCAGCAUUGGCAAUAUCCUCUCCGU ......---.((.(((((((....(((((...(((..(((((......)))))..)))..)))))...............((((.((((((....))))))..)))).))))))).)).. ( -43.30) >DroPse_CAF1 13400 117 + 1 CAUGUC---CGGCGAUGAUGAGGAGAUGCUGCGCAUCGCCUUCUUCAUCGGGGACUUUGUGCACCAGUACUACAUUCCGAUACUCUGCUGCACGGGCAGCAUCGGCAACAUCCUCUCCGU ......---(((.(((((.((((.(((((...))))).))))..)))))(((((....((((....))))......((((.....((((((....)))))))))).....))))).))). ( -41.60) >DroYak_CAF1 14695 117 + 1 UUCGGC---UGGCGAGGAUGAUGAGAUGCUAAGGAUAGCCUUCUUUAUAGGGGAUUUCGUGCACCAGUACUAUAUACCAGUGGUUUGCUGCACGGGCAGCAUUGGGAACAUCCUCUCCGU ......---.((.((((((((((((((((((....))))(((((....))))))))))))...((((((((((......)))))..(((((....))))))))))...))))))).)).. ( -40.50) >DroMoj_CAF1 15893 117 + 1 UAUGUC---CGUCGAGGAUGAGGAAAUGCUGCGAAUCGCGUACCUCAUUGGCGAUAUUGUGAAUAGAUAUUACGUGCCCAUCAUCUGCUGCACGGGCAGCAUUGGCAACAUACUCUCCGU ......---......(((.(((..((((((((..(((((...........))))).................(((((.((.....))..))))).))))))))(....)...)))))).. ( -32.60) >DroAna_CAF1 27682 120 + 1 CUCGGCGAGCGGCGAGGAUGAGGAGAUGCUGCGCAUCGCCUUUUUCAUCGGGGACGUUGUCCAUCAGUACUAUAUUCCCAUUCUGUGCUGCACCGGCAGCAUCGGCAACAUCCUCUCCGU ........((((.(((((((..(.((((((((((...)).........(((((((...))))..((((((..............))))))..)))))))))))..)..))))))).)))) ( -47.24) >DroPer_CAF1 13405 117 + 1 CAUGUC---CGGCGAUGAUGAGGAGAUGCUGCGCAUCGCCUUCUUCAUCGGGGACUUUGUGCACCAGUACUACAUUCCGAUACUCUGCUGCACGGGCAGCAUCGGCAACAUCCUCUCCGU ......---(((.(((((.((((.(((((...))))).))))..)))))(((((....((((....))))......((((.....((((((....)))))))))).....))))).))). ( -41.60) >consensus CACGGC___CGGCGAGGAUGAGGAGAUGCUGCGCAUCGCCUUCUUCAUCGGGGACUUUGUGCACCAGUACUACAUUCCAAUACUCUGCUGCACGGGCAGCAUCGGCAACAUCCUCUCCGU .........(((.(((((((....((((((((......((((.......))))...(((((((.(((.................))).))))))))))))))).....))))))).))). (-29.17 = -30.31 + 1.14)
| Location | 9,618,907 – 9,619,024 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -26.66 |
| Energy contribution | -28.46 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
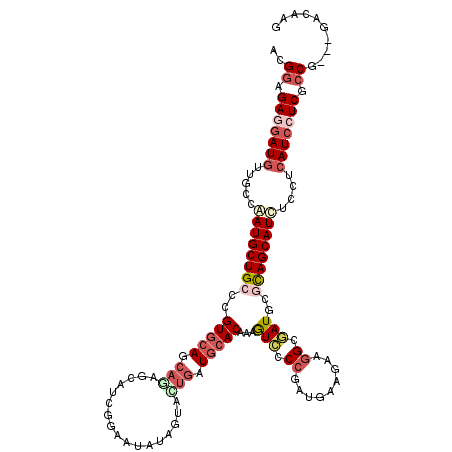
>3L_DroMel_CAF1 9618907 117 - 23771897 ACGGAGAGGAUAUUGCCAAUGCUGCCCGUGCAGCAAAGCACUGGUAUAUAGUAUUGAUGCACGAAGUGCCCUAUAAAAAAGGCUAUCCUUAGCAUCUCCUCAUCCUCGCCA---GCUGAU ..((.((((((......(((((((((.((((......)))).)))....))))))(((((...(((.(((..........)))....))).))))).....)))))).)).---...... ( -34.80) >DroPse_CAF1 13400 117 - 1 ACGGAGAGGAUGUUGCCGAUGCUGCCCGUGCAGCAGAGUAUCGGAAUGUAGUACUGGUGCACAAAGUCCCCGAUGAAGAAGGCGAUGCGCAGCAUCUCCUCAUCAUCGCCG---GACAUG .(((...((((.(((....((((((....))))))..(((((((.........))))))).))).)))).((((((.((.((.(((((...))))).)))).)))))))))---...... ( -42.60) >DroYak_CAF1 14695 117 - 1 ACGGAGAGGAUGUUCCCAAUGCUGCCCGUGCAGCAAACCACUGGUAUAUAGUACUGGUGCACGAAAUCCCCUAUAAAGAAGGCUAUCCUUAGCAUCUCAUCAUCCUCGCCA---GCCGAA .(((.((((((((((....((((((....))))))...(((..((((...))))..)))...)))............((..((((....))))...))..)))))))....---.))).. ( -36.80) >DroMoj_CAF1 15893 117 - 1 ACGGAGAGUAUGUUGCCAAUGCUGCCCGUGCAGCAGAUGAUGGGCACGUAAUAUCUAUUCACAAUAUCGCCAAUGAGGUACGCGAUUCGCAGCAUUUCCUCAUCCUCGACG---GACAUA ..((((((.(((((((...((((((((((.((.....))))))))).)))...............(((((...........)))))..)))))))...))).)))......---...... ( -32.50) >DroAna_CAF1 27682 120 - 1 ACGGAGAGGAUGUUGCCGAUGCUGCCGGUGCAGCACAGAAUGGGAAUAUAGUACUGAUGGACAACGUCCCCGAUGAAAAAGGCGAUGCGCAGCAUCUCCUCAUCCUCGCCGCUCGCCGAG .(((.(((((((((((((.((((((....))))))(((.((.........)).))).))).)))))))).(((.((...(((.(((((...))))).)))..)).)))....)).))).. ( -41.50) >DroPer_CAF1 13405 117 - 1 ACGGAGAGGAUGUUGCCGAUGCUGCCCGUGCAGCAGAGUAUCGGAAUGUAGUACUGGUGCACAAAGUCCCCGAUGAAGAAGGCGAUGCGCAGCAUCUCCUCAUCAUCGCCG---GACAUG .(((...((((.(((....((((((....))))))..(((((((.........))))))).))).)))).((((((.((.((.(((((...))))).)))).)))))))))---...... ( -42.60) >consensus ACGGAGAGGAUGUUGCCAAUGCUGCCCGUGCAGCAGAGCAUCGGAAUAUAGUACUGAUGCACAAAGUCCCCGAUGAAGAAGGCGAUGCGCAGCAUCUCCUCAUCCUCGCCG___GACAAG ..((.(((((((.....((((((((..(((((.(((.................))).)))))...(((.((.........)).)))..))))))))....))))))).)).......... (-26.66 = -28.46 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:45 2006