
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,616,146 – 9,616,314 |
| Length | 168 |
| Max. P | 0.763329 |

| Location | 9,616,146 – 9,616,251 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -44.68 |
| Consensus MFE | -31.25 |
| Energy contribution | -31.83 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
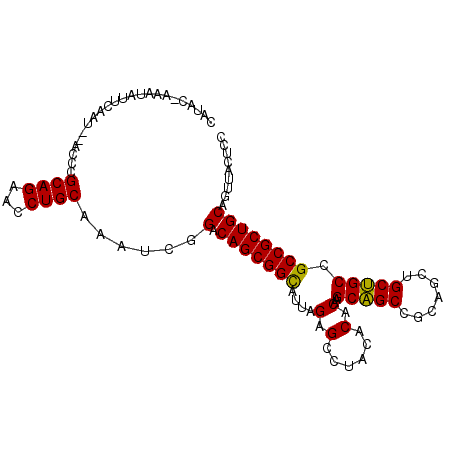
>3L_DroMel_CAF1 9616146 105 - 23771897 AAGCAGCCGCAGCUGCUGCCGCCGCUGCAGUUACUCCCGGCUCAAGUGGAAAUCGUCCACCUCGAGGCGCCGAUAAGAUUCGCGCCGCAUCGGGACCUUCCAUAC ..(((((.((.((....)).)).)))))......(((((((....(((((.....))))).....(((((.((......)))))))))..))))).......... ( -41.10) >DroVir_CAF1 12623 105 - 1 AGGCAGCUGCCGCUGCAGCCGCGGCUGCCGUGACGCCCAUUACGGGGAUCAGUCGCCCGCCUCGCGGCGCUGAUAAAAUUCGCGCUGCAUCGGGCCCCUCCAUAC .(((((((((.((....)).)))))))))..(((.(((......)))....)))(((((....(((((((.((......)))))))))..))))).......... ( -53.30) >DroPse_CAF1 10375 105 - 1 AAGCUGCAGCUGCCGCAGCCGCCGCUGCAGUCACGCCCGGUUCUGGUGGAAGUCGCCCGCCGCGCGGUGCAGACAAGAUACGCGCCGCCUCUGGUCCGUCUAUAC ..((((((((.((.......)).)))))))).(((.((((....(((((.......)))))..(((((((...........)))))))..))))..)))...... ( -41.30) >DroGri_CAF1 13131 105 - 1 AGGCAGCUGCUGCUGCAGCCGCUGCUGCUGUCACGCCCAUCACGGGCGGCAGUCGACCGCCGCGUGGCGCUGAUAAAAUACGUGCUGCAUCGGGUCCCUCGAUAC ..(((((.((....(((((....)))))(((((((((....(((((((((....).))))).))))))).)))))......)))))))((((((...)))))).. ( -44.10) >DroMoj_CAF1 12453 105 - 1 AGGCAGCCGCGGCUGCGGCUGCCGCUGCUGUUACACCCAUUACCGGAGGCAGUCGGCCGCCUCGGGGUGCUGAUAAAAUACGUGCUGCCUCGGGUCCGUCCAUAC .(((((((((....)))))))))..................(((.((((((((((((.(((....)))))))...........)))))))).))).......... ( -46.40) >DroPer_CAF1 10375 105 - 1 AAGCUGCAGCUGCCGCAGCCGCCGCUGCAGUCACGCCCGGUUCUGGUGGAAGUCGCCCGCCGCGCGGUGCAGACAAGAUACGGGCCGCCUCUGGUCCGUCUAUAC ..((((((((.((.......)).))))))))..((((.......))))...(((((((((...)))).)).))).....((((((((....))))))))...... ( -41.90) >consensus AAGCAGCAGCUGCUGCAGCCGCCGCUGCAGUCACGCCCAGUACGGGUGGAAGUCGCCCGCCGCGCGGCGCUGAUAAAAUACGCGCCGCAUCGGGUCCGUCCAUAC ..(((((.((.((....)).)).)))))......((((......(((((.......)))))..(((((((...........)))))))...)))).......... (-31.25 = -31.83 + 0.58)

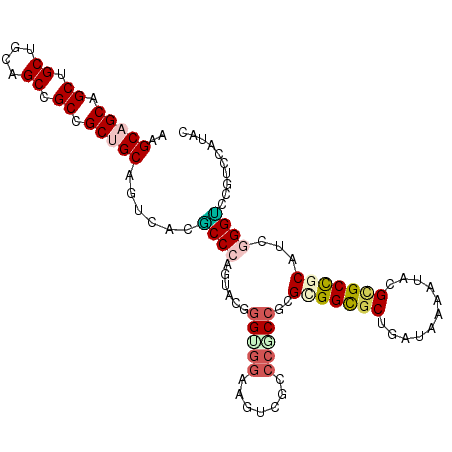
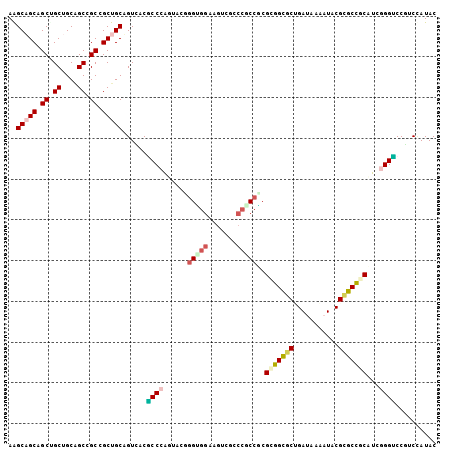
| Location | 9,616,214 – 9,616,314 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -28.54 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9616214 100 + 23771897 GGAGUAACUGCAGCGGCGGCAGCAGCUGCGGCUGCUUGUGUGUAGGCUCUAAUGCCGCUGUCCGAUUUGCAGGUUCUGCGGGA--AUUGAAUAUUGAAUAUA (.((...(((((.(((((((((((((....))))))........(((......))))))).)))...)))))...)).)....--................. ( -36.00) >DroPse_CAF1 10443 101 + 1 GGCGUGACUGCAGCGGCGGCUGCGGCAGCUGCAGCUUGGGUAUAGGCUCGAAUGCCGCUGUCCGACUUGCAGGUUCUGCGGGUGGGAUGAAUAUUA-GUUGG .....((((((((((((((((((((...)))))))((((((....)))))).))))))))(((.((((((((...)))))))).))).......))-))).. ( -46.80) >DroSec_CAF1 11863 99 + 1 GAAGUAACUGCAGCGGCGGCAGCAGCUGCGGCUGCUUGUGUGUAGGCUCUAAUACCGCUGUCGGAUUUGCAGGUUCUGCGGGA--AUUGAAUAUUU-AUAUA .(((((.(((((((((((((....)))))(.((((......)))).)........))))).))).(((((((...))))))).--......)))))-..... ( -31.10) >DroSim_CAF1 13898 99 + 1 GGAGUAACUGCAGCGGCGGCAGCAGCUGCGGCUGCUUGUGUGUAGGCUCUAAUGCCGCUGUCCGAUUUGCAGGUUCUGCGGGA--AUUGAAUAUUU-AUAUA (.((...(((((.(((((((((((((....))))))........(((......))))))).)))...)))))...)).)....--...........-..... ( -36.00) >DroYak_CAF1 11892 99 + 1 GGAGUAACUGCAGCGGCGGCAGCAGCUGCAGCUGCUUGGGUAUACGCCCUAAUGCCGCUGUCUGAUUUGCAGGUGCUGCGGAU--AACAAAUAUUU-GUAUG ..((((.(((((((((((((((((((....)))))..((((....))))...)))))))))......))))).))))((((((--(.....)))))-))... ( -42.70) >DroPer_CAF1 10443 101 + 1 GGCGUGACUGCAGCGGCGGCUGCGGCAGCUGCAGCUUGGGUAUAGGCUCGAAUGCCGCUGUCCGACUUGCAGGUUCUGCGGGUGGGAUGAACAUUA-GUUGG .....((((((((((((((((((((...)))))))((((((....)))))).))))))))(((.((((((((...)))))))).))).......))-))).. ( -46.80) >consensus GGAGUAACUGCAGCGGCGGCAGCAGCUGCGGCUGCUUGGGUAUAGGCUCUAAUGCCGCUGUCCGAUUUGCAGGUUCUGCGGGA__AAUGAAUAUUU_AUAUA .........((((((((((((((.......)))))).(.((....)).)....))))))))....(((((((...))))))).................... (-28.54 = -27.90 + -0.64)

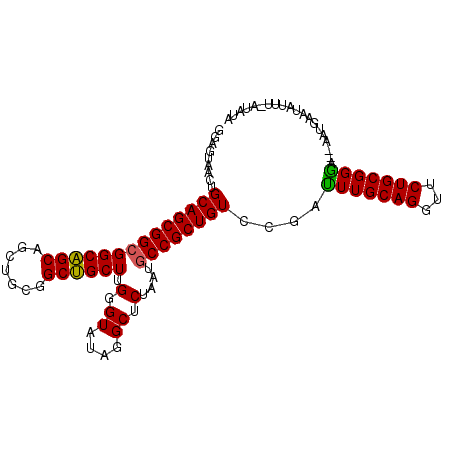
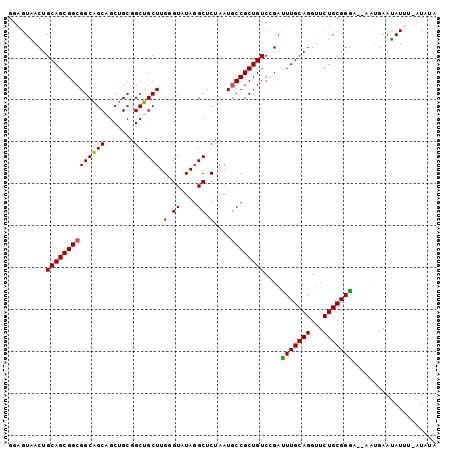
| Location | 9,616,214 – 9,616,314 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -24.17 |
| Energy contribution | -23.58 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9616214 100 - 23771897 UAUAUUCAAUAUUCAAU--UCCCGCAGAACCUGCAAAUCGGACAGCGGCAUUAGAGCCUACACACAAGCAGCCGCAGCUGCUGCCGCCGCUGCAGUUACUCC .................--....((((...))))......(.(((((((..(((...)))......((((((....))))))...))))))))......... ( -26.20) >DroPse_CAF1 10443 101 - 1 CCAAC-UAAUAUUCAUCCCACCCGCAGAACCUGCAAGUCGGACAGCGGCAUUCGAGCCUAUACCCAAGCUGCAGCUGCCGCAGCCGCCGCUGCAGUCACGCC .....-.........(((.((..((((...))))..)).)))..(((((......))).........((((((((.((.......)).))))))))...)). ( -29.20) >DroSec_CAF1 11863 99 - 1 UAUAU-AAAUAUUCAAU--UCCCGCAGAACCUGCAAAUCCGACAGCGGUAUUAGAGCCUACACACAAGCAGCCGCAGCUGCUGCCGCCGCUGCAGUUACUUC .....-...........--....((((...))))......(.(((((((..(((...)))......((((((....))))))...))))))))......... ( -24.10) >DroSim_CAF1 13898 99 - 1 UAUAU-AAAUAUUCAAU--UCCCGCAGAACCUGCAAAUCGGACAGCGGCAUUAGAGCCUACACACAAGCAGCCGCAGCUGCUGCCGCCGCUGCAGUUACUCC .....-...........--....((((...))))......(.(((((((..(((...)))......((((((....))))))...))))))))......... ( -26.20) >DroYak_CAF1 11892 99 - 1 CAUAC-AAAUAUUUGUU--AUCCGCAGCACCUGCAAAUCAGACAGCGGCAUUAGGGCGUAUACCCAAGCAGCUGCAGCUGCUGCCGCCGCUGCAGUUACUCC .....-...........--....(((((..(((.....)))...(((((....(((......))).((((((....))))))))))).)))))......... ( -35.40) >DroPer_CAF1 10443 101 - 1 CCAAC-UAAUGUUCAUCCCACCCGCAGAACCUGCAAGUCGGACAGCGGCAUUCGAGCCUAUACCCAAGCUGCAGCUGCCGCAGCCGCCGCUGCAGUCACGCC .....-.........(((.((..((((...))))..)).)))..(((((......))).........((((((((.((.......)).))))))))...)). ( -29.20) >consensus CAUAC_AAAUAUUCAAU__ACCCGCAGAACCUGCAAAUCGGACAGCGGCAUUAGAGCCUACACACAAGCAGCCGCAGCUGCUGCCGCCGCUGCAGUUACUCC .......................((((...))))......(.(((((((....(.(......).)..(((((.......))))).))))))))......... (-24.17 = -23.58 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:43 2006