
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,609,344 – 9,609,464 |
| Length | 120 |
| Max. P | 0.720630 |

| Location | 9,609,344 – 9,609,464 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -18.28 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
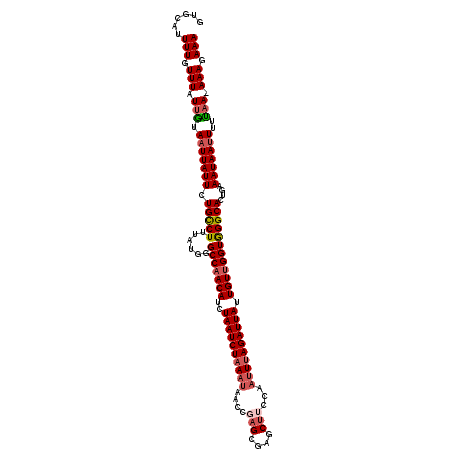
>3L_DroMel_CAF1 9609344 120 + 23771897 UUUCUUUUUUGGAAAUUAUUUAAGUGCCCACCAACAAUAAUCUUAAUUUGAAGCUCGCUCGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAGAAUAAUUACAAUAAACAAAAUGCAC (((.(((.(((..((((((((...((((..((((((.((((((....((((.(....)))))......))))))..))))))........)))))))))))).))).))).)))...... ( -20.30) >DroSec_CAF1 5061 119 + 1 UUUCUUU-UUAAAAAUUAUUUCAGUGCCCACCAACAAUAAUCUAAAUUAGAAGCUCGCUCGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGACAAAAUAAUUACAAUAAACAAAAUGCAC .......-...............((((...((((((.((((((((((..((.(....)))....))))))))))..))))))..(.....).........................)))) ( -18.00) >DroSim_CAF1 5068 119 + 1 UUUCUUU-UUAAAAAUUAUUUCAGUGCCCACCAACAAUAAUCUAAUUUAGAAGCUCGCUCGGUUAUUUAGAUUAGAUGUUGGCUCAUAAAGGCAAAAUAAUUACAAUAAACAAAAUGCAC .......-.....((((((((...((((..((((((....(((((((((((((((.....)))).)))))))))))))))))........)))))))))))).................. ( -22.40) >DroEre_CAF1 5033 119 + 1 UUUCUUU-UUAAAAAUUAUUUAAUUGCCUACCGACAAUAAUCUAAAGUGGUAGCUCGCGGGGGUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAGAAUAAUUAUAAUAAACAAAAUACAC .......-.....((((((((...(((((.((((((.(((((((((......((((....)))).)))))))))..)))))).......))))))))))))).................. ( -24.50) >DroYak_CAF1 5087 119 + 1 UUUCUUU-UGAAAAUUUAUUUGAUUGCCUACCAACAAUAAUCUAAAUUGGAAGCUCGCUGGGGUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAGAAUAAUUUUAAUAAACAAAAUACAC ......(-(((((..((((((...(((((.((((((.((((((((((.....((((....))))))))))))))..)))))).......))))))))))))))))).............. ( -24.90) >consensus UUUCUUU_UUAAAAAUUAUUUAAGUGCCCACCAACAAUAAUCUAAAUUAGAAGCUCGCUCGGUUAUUUAGAUUAGAUGUUGGCCCAUAAAGGCAGAAUAAUUACAAUAAACAAAAUGCAC .............((((((((...((((..((((((.((((((((((....((....)).....))))))))))..))))))........)))))))))))).................. (-18.28 = -19.08 + 0.80)



| Location | 9,609,344 – 9,609,464 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.92 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9609344 120 - 23771897 GUGCAUUUUGUUUAUUGUAAUUAUUCUGCCUUUAUGGGCCAACAUCUAAUCUAAAUAACCGAGCGAGCUUCAAAUUAAGAUUAUUGUUGGUGGGCACUUAAAUAAUUUCCAAAAAAGAAA ......(((.(((.(((.(((((((.(((((......(((((((..((((((.(((....(((....)))...))).)))))).))))))))))))....)))))))..))).))).))) ( -23.60) >DroSec_CAF1 5061 119 - 1 GUGCAUUUUGUUUAUUGUAAUUAUUUUGUCUUUAUGGGCCAACAUCUAAUCUAAAUAACCGAGCGAGCUUCUAAUUUAGAUUAUUGUUGGUGGGCACUGAAAUAAUUUUUAA-AAAGAAA .....((((.((((....(((((((((((((......(((((((..((((((((((....(((....)))...)))))))))).))))))))))))...))))))))..)))-).)))). ( -25.50) >DroSim_CAF1 5068 119 - 1 GUGCAUUUUGUUUAUUGUAAUUAUUUUGCCUUUAUGAGCCAACAUCUAAUCUAAAUAACCGAGCGAGCUUCUAAAUUAGAUUAUUGUUGGUGGGCACUGAAAUAAUUUUUAA-AAAGAAA .....((((.((((....(((((((((((((......(((((((..((((((((......(((....))).....)))))))).))))))))))))...))))))))..)))-).)))). ( -25.80) >DroEre_CAF1 5033 119 - 1 GUGUAUUUUGUUUAUUAUAAUUAUUCUGCCUUUAUGGGCCAACAUCUAAUCUAAAUACCCCCGCGAGCUACCACUUUAGAUUAUUGUCGGUAGGCAAUUAAAUAAUUUUUAA-AAAGAAA .....((((.((((....(((((((.(((((......(((.(((..(((((((((......(....).......))))))))).))).))))))))....)))))))..)))-).)))). ( -20.62) >DroYak_CAF1 5087 119 - 1 GUGUAUUUUGUUUAUUAAAAUUAUUCUGCCUUUAUGGGCCAACAUCUAAUCUAAAUACCCCAGCGAGCUUCCAAUUUAGAUUAUUGUUGGUAGGCAAUCAAAUAAAUUUUCA-AAAGAAA .....(((((......(((.(((((.(((((......(((((((..((((((((((.....((....))....)))))))))).))))))))))))....))))).))).))-))).... ( -23.00) >consensus GUGCAUUUUGUUUAUUGUAAUUAUUCUGCCUUUAUGGGCCAACAUCUAAUCUAAAUAACCGAGCGAGCUUCCAAUUUAGAUUAUUGUUGGUGGGCACUGAAAUAAUUUUUAA_AAAGAAA ......(((.(((.(((.(((((((.(((((......(((((((..((((((((((....(((....)))...)))))))))).))))))))))))....)))))))..))).))).))) (-20.92 = -21.92 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:39 2006