
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,596,355 – 9,596,464 |
| Length | 109 |
| Max. P | 0.721722 |

| Location | 9,596,355 – 9,596,464 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.03 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -18.87 |
| Energy contribution | -18.74 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9596355 109 - 23771897 UAUUGUCGCCCUGUUCCACUGCCUUGGUAGCCUUGUCGUGGGCAGCCUUGGCUCGCUCUAGCAGCUCGAUGUCCUCCAGUUGCUGCUGUUCAGCUCUGAAAU------UAUAGAU----- ....((((..(((((.....(((..(((.((((......)))).)))..))).......)))))..))))......(((..((((.....)))).)))....------.......----- ( -32.10) >DroVir_CAF1 18934 110 - 1 UACUGUCGCCCUGCUGUACCGCCUUGGUGGCGCGUUCGUGGGCAGCCUUGGCGCGCUCCAAGAGCUCGAUGUCAUCCUUCUUUUGUGCAUCGGCGCUGG-GU------A---GUAAAAUU (((((..((((.((...((((((.....)))).))..))))))...((..((((((((...)))).((((((((.........)).))))))))))..)-))------)---)))..... ( -42.50) >DroGri_CAF1 19231 111 - 1 UAUUGUCGCCCUGCUUCACCGCCUUGGUAGCCUUAUCAAGGGCAGCCUUGGCGCGUUCCAACAAUUCAAUGUCAUCCUUCUUUUGUGCAUCGGCGCUUAAGG------A---AGAAAUAU (((((......((....((((((..(((.(((((....))))).)))..)))).))..))......))))).....(((((((.((((....))))..))))------)---))...... ( -32.60) >DroEre_CAF1 21839 107 - 1 UAUUGUCGCCCUGUUCCACUGCCUUGGUGGCCUUGUCGUGGGCGGCCUUAGCUCGCUCCAAGAGCUCGAUGUCUUCCAGUUGCUGCUGCUCAGCUCUGUAAU------GUA--AU----- ....(((((((.....((..(((.....)))..))....)))))))...(((((.......)))))..........(((..((((.....)))).)))....------...--..----- ( -30.90) >DroWil_CAF1 21480 115 - 1 UAUUAUCACCCUGUUUUACAGCCUUGGUGGCCUUCUCGAAGGCUGCCUUGGCCCGCUCCAAGAGCUCAAUGUCAUCUUGUUUUUGUUUUGCGGCCCUGAAACAGACAUAAUUGUU----- ..........((((((((..(((..((..(((((....)))))..))..)))((((..(((((((.............)))))))....))))...))))))))...........----- ( -39.22) >DroAna_CAF1 23313 108 - 1 UAUUGUCUCCUUGCUCCACGGCCUCGGUGGCCUUGUCGUGAGCAGCCUUGGCUCUCGCCAGUAGCUCAAUGUCCUCCUGCUGCUGCUUGUCGGCUCUA-AAU------AAUACGG----- ((((((.............((((.....))))..((((((((((((.(((((....))))).(((.............))))))))))).))))....-.))------))))...----- ( -32.02) >consensus UAUUGUCGCCCUGCUCCACCGCCUUGGUGGCCUUGUCGUGGGCAGCCUUGGCUCGCUCCAAGAGCUCAAUGUCAUCCUGCUGCUGCUCAUCGGCUCUGAAAU______AAU_GAU_____ ............(((.....(((..(((.((((......)))).)))..)))......(((((((.............)))))))......))).......................... (-18.87 = -18.74 + -0.14)

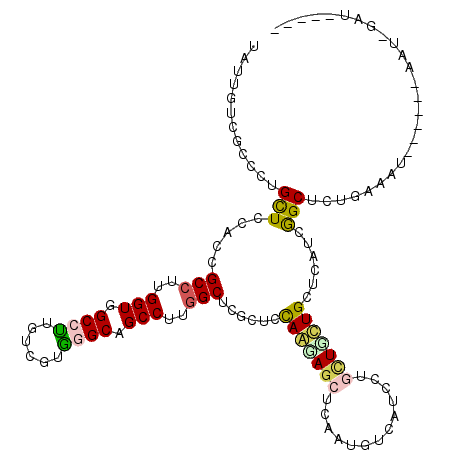
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:28 2006