
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
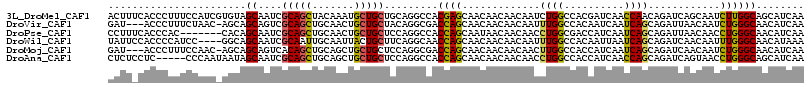
| Location | 9,566,649 – 9,566,765 |
| Length | 116 |
| Max. P | 0.978522 |

| Location | 9,566,649 – 9,566,765 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.11 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
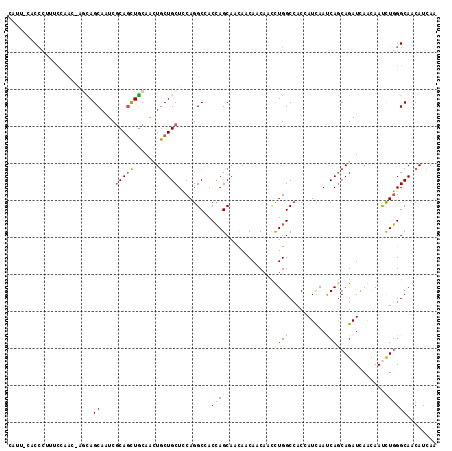
>3L_DroMel_CAF1 9566649 116 + 23771897 ACUUUCACCCUUUCCAUCGUGUAGCAAUCGCAGCUACAAAUGCUGCUGCAGGCCACGAGCAACAACAACAAUCUGGCCACGAUCAACCAACAGAUCAGCAAUCUUGGCAGCAUCAA ...................((((((.......)))))).((((((((((.(((((.((.............)))))))..((((........)))).))......))))))))... ( -32.02) >DroVir_CAF1 1441 112 + 1 GAU---ACCCUUUCUAAC-AGCAGCAGUCGCAGCUGCAACUGCUGCUACAGGCGACCAGCAACAACAACAAUUUGGCCACAAUCAAUCAGCAGAUUAACAAUCUGGGCAACAUCAA (((---............-(((((((((.((....)).)))))))))...(((......................)))...)))......(((((.....)))))(....)..... ( -27.35) >DroPse_CAF1 2813 109 + 1 CCUUUCACCCAC-------CACAGCAAUCGCAGCUGCAACUGCUGCUCCAGGCCACCAGCAAUAACAACAACCUGGCGACCAUCAAUCAGCAGAUUAACAACCUGGGCAACAUCAA .......((((.-------..((((.......))))...((((((.....((.(.((((.............)))).).))......))))))..........))))......... ( -24.22) >DroWil_CAF1 1261 112 + 1 UAUUCCACCCCAUCC----GGCAGCAAUCGCAAUUGCAAUUACUGCUUCAGGCAACCAGCAACAACAACAAUUUGGCCACAAUUAAUCAGCAGAUCAACAAUUUGGGCAACAUAAA ........((((...----(((.(((((....)))))......((((...(....).))))..............))).......(((....)))........))))......... ( -18.10) >DroMoj_CAF1 1493 112 + 1 GAU---ACCCUUUCCAAC-AGCAGCAGUCACAGCUGCAGCUGCUGCUCCAGGCGACCAGCAACAACAACAACUUGGCCACCAUCAAUCAGCAGAUCAACAAUCUGGGCAACAUCAA (((---.(((........-.(((((.......)))))..((((((.....((.(.((((.............)))).).))......))))))...........)))....))).. ( -25.62) >DroAna_CAF1 1608 111 + 1 CUCUCCUC-----CCCAAUAAUAGCAAUCGCAGCUGCAGCUGCUGCUCCAGGCCACCAGCAACAACAACAACCUGGCCACCAUCAACCAGCAGAUCAGUAACCUGGGCAGCAUCAA ........-----..........((....)).(((((..((((((.....(((((..................))))).........)))))).((((....)))))))))..... ( -26.31) >consensus CAUU_CACCCUUUCCAAC_AGCAGCAAUCGCAGCUGCAACUGCUGCUCCAGGCCACCAGCAACAACAACAACCUGGCCACCAUCAAUCAGCAGAUCAACAAUCUGGGCAACAUCAA .......................((....(((((.......))))).........((((.............((((..........))))............))))))........ (-12.69 = -12.11 + -0.58)

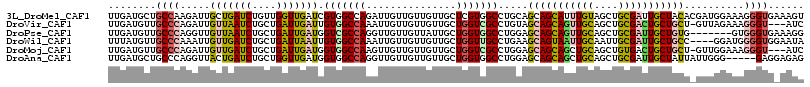
| Location | 9,566,649 – 9,566,765 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
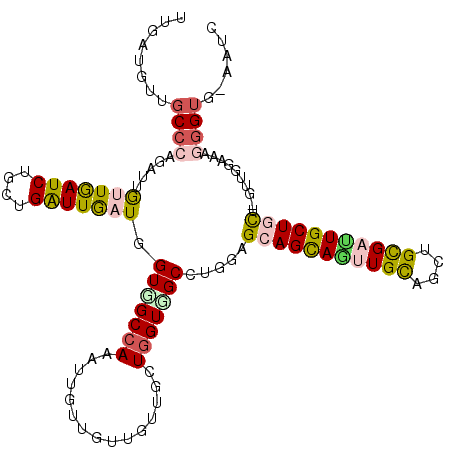
>3L_DroMel_CAF1 9566649 116 - 23771897 UUGAUGCUGCCAAGAUUGCUGAUCUGUUGGUUGAUCGUGGCCAGAUUGUUGUUGUUGCUCGUGGCCUGCAGCAGCAUUUGUAGCUGCGAUUGCUACACGAUGGAAAGGGUGAAAGU ...............(..((..(((((((((.(((((..(((((((.((((((((.((.....))..)))))))))))))..))..)))))...)).)))))))...))..).... ( -35.50) >DroVir_CAF1 1441 112 - 1 UUGAUGUUGCCCAGAUUGUUAAUCUGCUGAUUGAUUGUGGCCAAAUUGUUGUUGUUGCUGGUCGCCUGUAGCAGCAGUUGCAGCUGCGACUGCUGCU-GUUAGAAAGGGU---AUC .......(((((.....(((((((....))))))).(((((((...............)))))))...(((((((((((((....))))))))))))-).......))))---).. ( -42.36) >DroPse_CAF1 2813 109 - 1 UUGAUGUUGCCCAGGUUGUUAAUCUGCUGAUUGAUGGUCGCCAGGUUGUUGUUAUUGCUGGUGGCCUGGAGCAGCAGUUGCAGCUGCGAUUGCUGUG-------GUGGGUGAAAGG ......(..((((((..(((((((....)))))))..)).((((((..((((....)).))..)))))).(((((((((((....))))))))))).-------.))))..).... ( -47.60) >DroWil_CAF1 1261 112 - 1 UUUAUGUUGCCCAAAUUGUUGAUCUGCUGAUUAAUUGUGGCCAAAUUGUUGUUGUUGCUGGUUGCCUGAAGCAGUAAUUGCAAUUGCGAUUGCUGCC----GGAUGGGGUGGAAUA ......(..(((.....(((((((....))))))).(..((((...............))))..)(((..(((((((((((....))))))))))))----))...)))..).... ( -35.66) >DroMoj_CAF1 1493 112 - 1 UUGAUGUUGCCCAGAUUGUUGAUCUGCUGAUUGAUGGUGGCCAAGUUGUUGUUGUUGCUGGUCGCCUGGAGCAGCAGCUGCAGCUGUGACUGCUGCU-GUUGGAAAGGGU---AUC .......(((((.....((..(((....)))..))((((((((.((..........)))))))))).....(((((((.((((......)))).)))-))))....))))---).. ( -42.00) >DroAna_CAF1 1608 111 - 1 UUGAUGCUGCCCAGGUUACUGAUCUGCUGGUUGAUGGUGGCCAGGUUGUUGUUGUUGCUGGUGGCCUGGAGCAGCAGCUGCAGCUGCGAUUGCUAUUAUUGGG-----GAGGAGAG ....(.((.(((((((((((.(((........)))))))))).((((((.(((((.((((.((........)).)))).))))).))))))........))))-----.)).)... ( -39.50) >consensus UUGAUGUUGCCCAGAUUGUUGAUCUGCUGAUUGAUGGUGGCCAAAUUGUUGUUGUUGCUGGUGGCCUGGAGCAGCAGUUGCAGCUGCGAUUGCUGCU_GUUGGAAAGGGUG_AAUC ........((((.....(((((((....))))))).(((((((...............))))))).....(((((((((((....)))))))))))..........))))...... (-26.10 = -26.58 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:21 2006