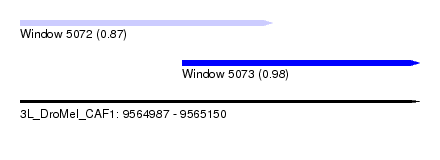
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,564,987 – 9,565,150 |
| Length | 163 |
| Max. P | 0.980251 |

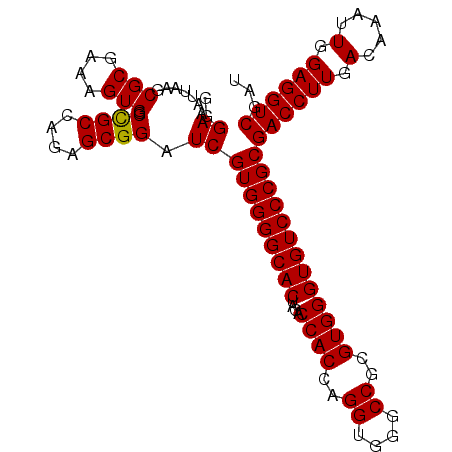
| Location | 9,564,987 – 9,565,090 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -45.03 |
| Consensus MFE | -41.85 |
| Energy contribution | -41.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9564987 103 + 23771897 GGGAAAUUAAGCGCGAAAGUGGCUGCCAGUGCGGAUCGUGGGGCACUAGACCACCAGGUGGGCCGCGUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAU ((((.....(((((....)).)))(((..(((((..(.((((((....).)).))).)....)))))..))).)))).(((((((.(.....).))))))).. ( -42.10) >DroSim_CAF1 3584 103 + 1 GGGAAAUUAAGCGCGAAAGUGGCCGCCAGAGCGGAUCGUGGGGCACUAGACCACCAGGUGGGCCGUGUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAU ...........(((....))).((((....))))((((((((((((....((((..((....))..)))))))))))))((((((.(.....).))))))))) ( -43.30) >DroYak_CAF1 558 103 + 1 GGGAAAUUAAGCGCGAAAGUGGCCGCCAGCGCGGAUCGUGGGGCACUAGACCACCAGGUGGGCCGCGUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGCU ((((.......(((....)))..((((.((((((..(.((((((....).)).))).)....)))))).))))))))((((((((.(.....).)))))))). ( -49.70) >consensus GGGAAAUUAAGCGCGAAAGUGGCCGCCAGAGCGGAUCGUGGGGCACUAGACCACCAGGUGGGCCGCGUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAU ..((.......(((....))).((((....)))).))(((((((((....((((..((....))..)))))))))))))((((((.(.....).))))))... (-41.85 = -41.63 + -0.22)


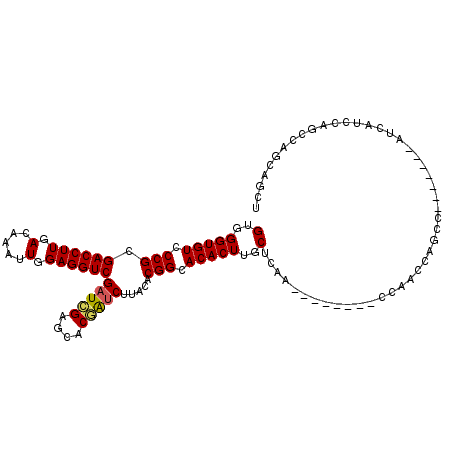
| Location | 9,565,053 – 9,565,150 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.52 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9565053 97 + 23771897 GUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAUUGAGCACAAUCUUACACGGCACACUUGCUCAA--------CCAACCAGCC-------AUCAUCCAGCCAGCAGCU (..(((((.(((.((((((.(.....).))))))(((((....))))).....))).)))))..)....--------..........-------.......(((.....))) ( -27.10) >DroSim_CAF1 3650 112 + 1 GUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAUCGAGCACGGUCUUACACGGCACACUUGCUCAACAAACGAACCAACCAGCCAGCCAUCAUCAUCCAGCCAGCAGCA (..(((((.(((.((((((.(.....).))))))(((((....))))).....))).)))))..)...................((..((............))..)).... ( -29.50) >DroYak_CAF1 624 86 + 1 GUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGCUGGUUUUCGAACUUACACGGCACACUUGCUUAA--------CCA------------------CCUACCAAACAGCU (((((((.((.((((((((.(.....).)))))))).))...............((((....))))...--------.))------------------)))))......... ( -27.80) >consensus GUGGGUGUCCCGCGACCUUGACAAAUUGGAGGUCGAUCGAGCACGAUCUUACACGGCACACUUGCUCAA________CCAACCAGCC_______AUCAUCCAGCCAGCAGCU (..(((((.(((.((((((.(.....).))))))(((((....))))).....))).)))))..)............................................... (-24.54 = -24.77 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:19 2006