| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,545,710 – 9,545,848 |
| Length | 138 |
| Max. P | 0.801602 |

| Location | 9,545,710 – 9,545,817 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9545710 107 + 23771897 CGUGUAAAGCAAAUGUUAAACAUGAAUGUGUGUAAGGGAAUAUGUGCAAGGGGUUACUGAAAUGUACUUUUCACAUUUCAUAAUUUAUUACUACACACUAAAAAUUC .(((((..(((.(((((..((((......)))).....))))).)))...((((((.((((((((.......)))))))))))))).....)))))........... ( -21.30) >DroSec_CAF1 54708 107 + 1 AGUGUAAAGCAAAUGUUAAACAUGAAAGUGUGUAAGUGAAUAUGUGCAUGGGGUUACUGAAAUGUACUUUUCACAUUUCAUAAUUUAUUACUACACAUUAAAAAUUC .(((((..(((.(((((..((((....)))).......))))).)))...((((((.((((((((.......)))))))))))))).....)))))........... ( -23.00) >DroSim_CAF1 56807 107 + 1 AGUGUAAAGCAAAUGUUAAACAUGAAAGUGUGUAAGUGAAUAUGUGCAUGGGGUUACUGAAAUGUACUUUUCACAUUUCAUAAUUUAUUACUACACAUUAAAAAUUC .(((((..(((.(((((..((((....)))).......))))).)))...((((((.((((((((.......)))))))))))))).....)))))........... ( -23.00) >DroEre_CAF1 55018 107 + 1 AGUGUAAAGCAAAUGUUGAACAUGAAAGUGUGUGUGUGAAUAUGUACAAGGGGUUACUGAAAUGCACUUUCCACAUUUCAUAAUUUAUUACUGUACAUUCAAAAUUC ((((((.((.((((..(((((((....))))(((((.(((..((((...((.....))....))))..)))))))).)))..))))....)).))))))........ ( -18.40) >DroYak_CAF1 58166 107 + 1 AGUGUAAAGCAUAUGUUAAACAUGCAGGUGUGUAAGUGAAUAUGUGCAAGGGGUUAGUGAAAUGUUCUUUUCACAUUUCAUAAUUUAUUACUAUACAUUAAAAAUUC ((((((..(((((((((..((.((((....)))).)).)))))))))...((((((.((((((((.......)))))))))))))).......))))))........ ( -24.50) >consensus AGUGUAAAGCAAAUGUUAAACAUGAAAGUGUGUAAGUGAAUAUGUGCAAGGGGUUACUGAAAUGUACUUUUCACAUUUCAUAAUUUAUUACUACACAUUAAAAAUUC .(((((..(((.(((((..((((......)))).....))))).)))...((((((.((((((((.......)))))))))))))).....)))))........... (-18.22 = -18.22 + 0.00)

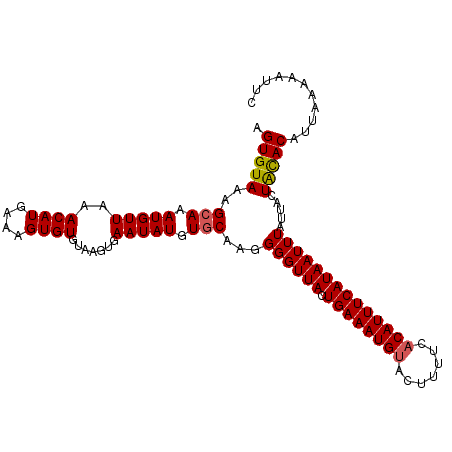
| Location | 9,545,710 – 9,545,817 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -16.89 |
| Consensus MFE | -13.99 |
| Energy contribution | -13.67 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9545710 107 - 23771897 GAAUUUUUAGUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCUUGCACAUAUUCCCUUACACACAUUCAUGUUUAACAUUUGCUUUACACG .........((((((((..........(((((((((.....)))))))))((((....))))..........))))))))........................... ( -19.10) >DroSec_CAF1 54708 107 - 1 GAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCAUGCACAUAUUCACUUACACACUUUCAUGUUUAACAUUUGCUUUACACU ((((.....(((((((...........(((((((((.....))))))))).........)))))))))))..................................... ( -16.05) >DroSim_CAF1 56807 107 - 1 GAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCAUGCACAUAUUCACUUACACACUUUCAUGUUUAACAUUUGCUUUACACU ((((.....(((((((...........(((((((((.....))))))))).........)))))))))))..................................... ( -16.05) >DroEre_CAF1 55018 107 - 1 GAAUUUUGAAUGUACAGUAAUAAAUUAUGAAAUGUGGAAAGUGCAUUUCAGUAACCCCUUGUACAUAUUCACACACACACUUUCAUGUUCAACAUUUGCUUUACACU ..........((((.(((((.....((((((((((((((.(((((..............)))))...))).)))).....)))))))........))))).)))).. ( -17.66) >DroYak_CAF1 58166 107 - 1 GAAUUUUUAAUGUAUAGUAAUAAAUUAUGAAAUGUGAAAAGAACAUUUCACUAACCCCUUGCACAUAUUCACUUACACACCUGCAUGUUUAACAUAUGCUUUACACU ..........((((..((((....(((((((((((.......)))))))).)))....))))....................((((((.....))))))..)))).. ( -15.60) >consensus GAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCUUGCACAUAUUCACUUACACACUUUCAUGUUUAACAUUUGCUUUACACU ((((.....(((((((...........(((((((((.....))))))))).........)))))))))))..................................... (-13.99 = -13.67 + -0.32)

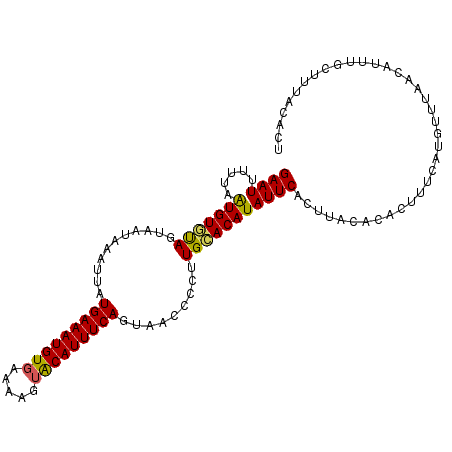
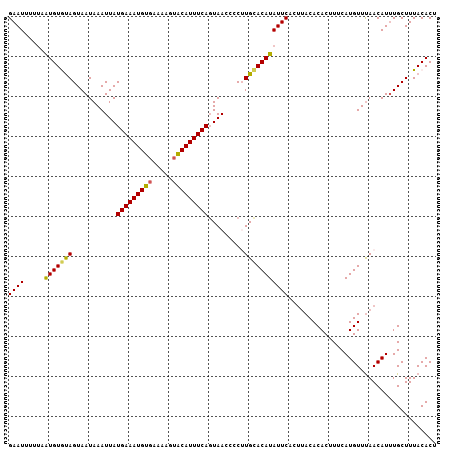
| Location | 9,545,739 – 9,545,848 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
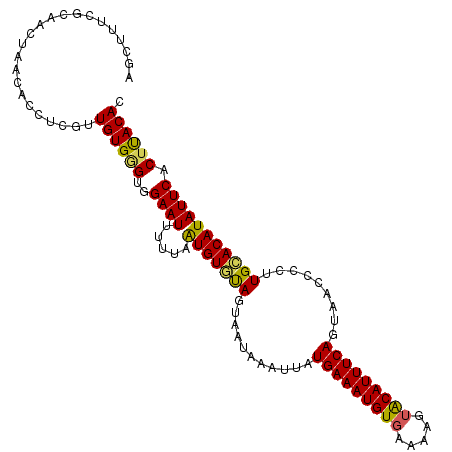
>3L_DroMel_CAF1 9545739 109 - 23771897 AGCUUUCGGUGCUAACACCUCGUUGUGAGUGGAAUUUUUAGUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCUUGCACAUAUUCCCUUACAC .......((((....))))....((((((.(((((.....((((((((..........(((((((((.....)))))))))........))))))))))))))))))). ( -30.57) >DroSec_CAF1 54737 109 - 1 AGCUUUCGCUGCUAACACCUCUUUGUGGGUGGAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCAUGCACAUAUUCACUUACAC (((.......)))..........((((((((((.......(((((((...........(((((((((.....))))))))).........))))))).)))))))))). ( -25.25) >DroSim_CAF1 56836 102 - 1 AGCUUUCG-------CACCUCGUUGUGGGUGGAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCAUGCACAUAUUCACUUACAC .((....)-------).......((((((((((.......(((((((...........(((((((((.....))))))))).........))))))).)))))))))). ( -24.55) >DroEre_CAF1 55047 109 - 1 AGCUUUCGUAACUAACACCUCGUUGUGGGCGGAAUUUUGAAUGUACAGUAAUAAAUUAUGAAAUGUGGAAAGUGCAUUUCAGUAACCCCUUGUACAUAUUCACACACAC ...((((((...((((.....))))...))))))...(((((((((((..........((((((((.......))))))))........))))))))..)))....... ( -21.37) >DroYak_CAF1 58195 109 - 1 AGCUUUCGCAACUAACACCUCGUUGUGGGCGGAAUUUUUAAUGUAUAGUAAUAAAUUAUGAAAUGUGAAAAGAACAUUUCACUAACCCCUUGCACAUAUUCACUUACAC .((...((((((.........))))))((.((......((((.(((....))).))))((((((((.......))))))))....))))..))................ ( -18.40) >consensus AGCUUUCGCAACUAACACCUCGUUGUGGGUGGAAUUUUUAAUGUGUAGUAAUAAAUUAUGAAAUGUGAAAAGUACAUUUCAGUAACCCCUUGCACAUAUUCACUUACAC .......................((((((..((((.....(((((((...........(((((((((.....))))))))).........))))))))))).)))))). (-17.47 = -17.07 + -0.40)

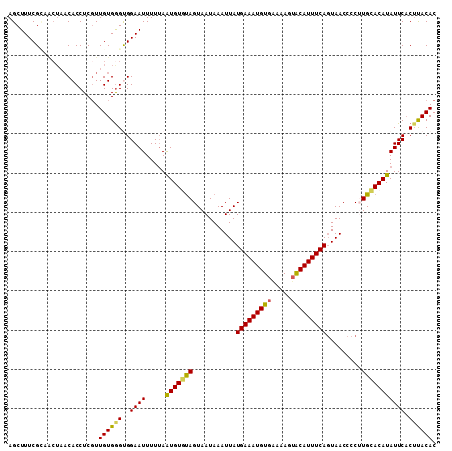
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:16 2006