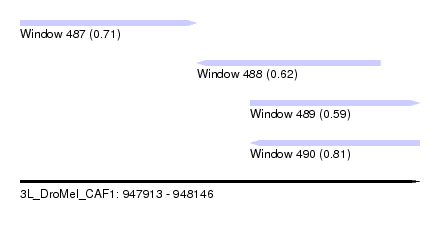
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 947,913 – 948,146 |
| Length | 233 |
| Max. P | 0.813104 |

| Location | 947,913 – 948,016 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.39 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 947913 103 + 23771897 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGUG----------------GAAUGGUGCUGCGGAUCCUGAAAGGGG-AAGGA ..((((((((.(((((........)))))))))))))...((.(((..(((........))).(((((((.----------------.......)))))))..(((....))))-)))). ( -37.70) >DroSec_CAF1 82941 103 + 1 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAAUG----------------GAAUGGGGCUGCGGAUCCUGAAAGGGG-AAGGA ..((((((((.(((((........)))))))))))))...((.(((..(((........))).(((((...----------------.........)))))..(((....))))-)))). ( -31.80) >DroSim_CAF1 74856 103 + 1 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGUG----------------GAAUGGGGCUGCGGAUCCUGAAAGGGG-AAGGA ..((((((((.(((((........)))))))))))))...((.(((..(((........))).(((((((.----------------.......)))))))..(((....))))-)))). ( -37.30) >DroEre_CAF1 84898 104 + 1 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGUG----------------GAAUGGGGCGGCGGGUCCUGAAAGGGGAAAUGA ..((((((((.(((((........)))))))))))))...((.((((.((((......((((.((.((...----------------...)).)).)))))))).)))).))........ ( -33.30) >DroYak_CAF1 87126 111 + 1 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGUGAA---------AGGGGGAAGGGGGCUGCGGAUCCUGAAAGGGGCAAGGA ..((((((((.(((((........)))))))))))))...((......(((........))).(((((((...---------............))))))).((((....))))...)). ( -34.96) >DroAna_CAF1 89363 120 + 1 AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGAGUGGGCCGCAUAGAGACGAGGCGUGGUGCAGGACGUGGGACGUGGGAGGA ..((((((((.(((((........)))))))))))))...((.((((...((((((.(..(((((((...))))(((.(........).))).)))..)....))))))...)))).)). ( -37.10) >consensus AUGUGUCGAAUGUUACGCUAAGGAGUAACUUUGACAUUUACCAUUCAAGGCUUAUGAGUGCCACCGCAGUG________________GAAUGGGGCUGCGGAUCCUGAAAGGGG_AAGGA ..((((((((.(((((........)))))))))))))...((.((((.(((........))).(((((((........................)))))))....)))).))........ (-28.67 = -29.39 + 0.72)

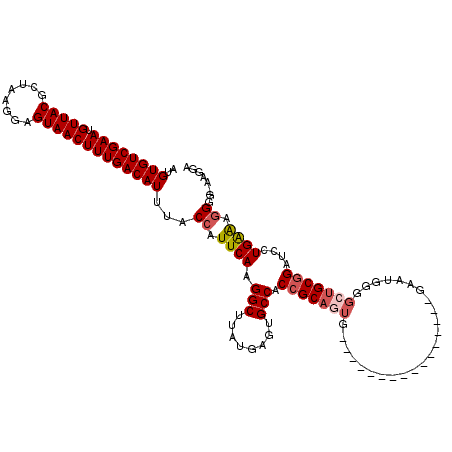
| Location | 948,016 – 948,123 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -34.69 |
| Energy contribution | -35.22 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 948016 107 - 23771897 ----GUCCUCAUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGGCA---------ACACGGUA ----((((......(((((((((..(((((..(....)....)))))..)))))))))(((.(((((((....))).((....)).....)))).))))))).---------........ ( -40.80) >DroSec_CAF1 83044 107 - 1 ----GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGGCA---------ACACGGUA ----((((......(((((((((..(((((..(....)....)))))..)))))))))(((.(((((((....))).((....)).....)))).))))))).---------........ ( -40.80) >DroSim_CAF1 74959 107 - 1 ----GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGGCA---------ACACGGUA ----((((......(((((((((..(((((..(....)....)))))..)))))))))(((.(((((((....))).((....)).....)))).))))))).---------........ ( -40.80) >DroEre_CAF1 85002 102 - 1 ----GUCCU-----CUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGCUU---------GCACGGUA ----(((..-----(((((((((..(((((..(....)....)))))..)))))))))(((.(((((((....))).((....)).....)))).))))))..---------........ ( -36.40) >DroYak_CAF1 87237 116 - 1 ----GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCAGGCGGGUUGCAAGCCGUGCACGGUA ----(((((....((((((((((..(((((..(....)....)))))..))))))))))..)))))((((........))))..((((((((....)).))))))..((((....)))). ( -42.80) >DroAna_CAF1 89483 109 - 1 UCACCUUCUUGUCCCUGCCCAUGUCCGUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGGUU---------AC--AGCA ........((((.(((((((..............((((((.(((......)))))))))...(((((((....))).((....)).....)))))))))))..---------))--)).. ( -37.10) >consensus ____GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCAUAAGGCAAUUGCCCGGGCGGGCA_________ACACGGUA ....((((......(((((((((..(((((..(....)....)))))..)))))))))(((.(((((((....))).((....)).....)))).))))))).................. (-34.69 = -35.22 + 0.53)

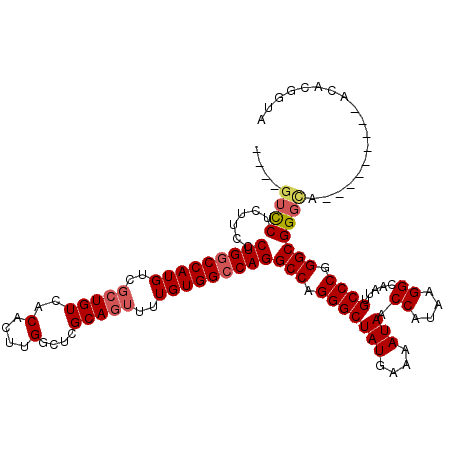
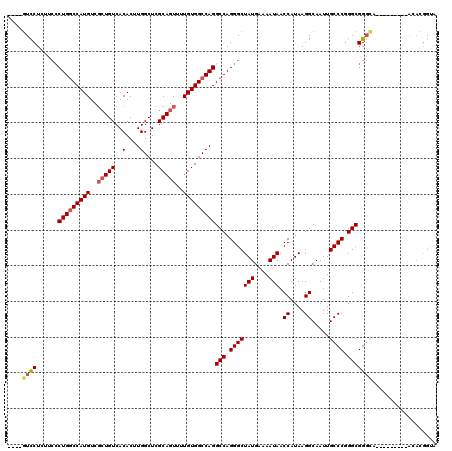
| Location | 948,047 – 948,146 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 948047 99 + 23771897 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAGGGAUGAGGAC----------CCAGCAG-AGGACCU------GGUA----ACUUG .((((((((((((..(((((((((((((.((....)).).)))).((((.......))))))))))))))))))).----------((((...-.....))------))))----))).. ( -35.00) >DroSec_CAF1 83075 99 + 1 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAGGGAAGAGGAC----------CAGGCAA-AGGACCU------GGUA----ACUUG .((((((((((....(((((((((((((.((....)).).)))).((((.......))))))))))))..)))))(----------(((((..-..).)))------))))----))).. ( -37.00) >DroSim_CAF1 74990 99 + 1 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAGGGAAGAGGAC----------CAGGCAA-AGGACCU------GGUA----ACUUG .((((((((((....(((((((((((((.((....)).).)))).((((.......))))))))))))..)))))(----------(((((..-..).)))------))))----))).. ( -37.00) >DroEre_CAF1 85033 82 + 1 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAG-----AGGAC----------CUGGCA-----------------------ACUUG .((((((....))))))(((((((((((.((....)).).)))).((((.......))))))))))-----.....----------..(...-----------------------.)... ( -23.90) >DroYak_CAF1 87277 104 + 1 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAGGGAAGAGGAC----------CAAGCAAGAGGACCA------GCCAAAGGACUUG (((((..........(((((((((((((.((....)).).)))).((((.......))))))))))))....((.(----------(........)).)))------))))......... ( -34.10) >DroAna_CAF1 89512 118 + 1 UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACACGGACAUGGGCAGGGACAAGAAGGUGAAGAUGGCUGGCAAGGAGACGAAAAGGAGGC--AGGACUUG .((((((((((((..(((((.(((((((.((....)).).)))).((((.......)))))).)))))........)))))))))))).....(....)......(((..--....))). ( -34.90) >consensus UGGUUAUUUUCAUAGCCCUGGCCUGGCCACAAAACUGCGAGCCAAGUGUGACAGCGACAUGGCCAGGGAAGAGGAC__________CAGGCAA_AGGACCU______GGUA____ACUUG ........(((....(((((((((((((.((....)).).)))).((((.......))))))))))))....)))............................................. (-23.52 = -23.88 + 0.36)

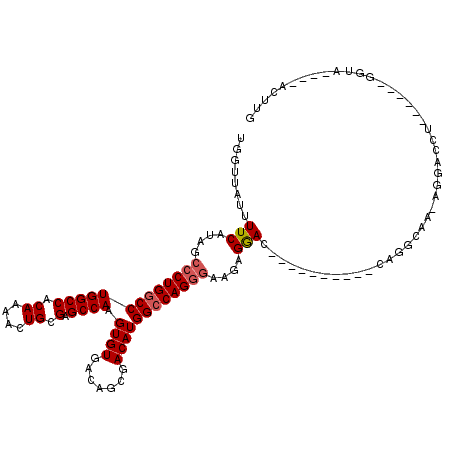
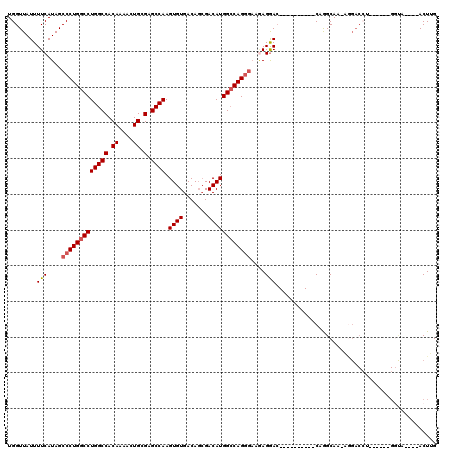
| Location | 948,047 – 948,146 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 948047 99 - 23771897 CAAGU----UACC------AGGUCCU-CUGCUGG----------GUCCUCAUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA ...((----((..------.((.(((-.....))----------).))((((((((((((.(((.......)))..((((.(((......)))))))))))))))..))))...)))).. ( -35.00) >DroSec_CAF1 83075 99 - 1 CAAGU----UACC------AGGUCCU-UUGCCUG----------GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA .(((.----.(((------((((...-..)))))----------))...)))((((((((.(((.......)))..((((.(((......))))))))))))))).(((....))).... ( -37.10) >DroSim_CAF1 74990 99 - 1 CAAGU----UACC------AGGUCCU-UUGCCUG----------GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA .(((.----.(((------((((...-..)))))----------))...)))((((((((.(((.......)))..((((.(((......))))))))))))))).(((....))).... ( -37.10) >DroEre_CAF1 85033 82 - 1 CAAGU-----------------------UGCCAG----------GUCCU-----CUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA ...((-----------------------(..(((----------(((((-----(((((((((..(((((..(....)....)))))..)))))))))...)))))).))..)))..... ( -27.20) >DroYak_CAF1 87277 104 - 1 CAAGUCCUUUGGC------UGGUCCUCUUGCUUG----------GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA ..((((((..(((------.((.((........)----------).))......(((((((((..(((((..(....)....)))))..))))))))))))))))))............. ( -37.00) >DroAna_CAF1 89512 118 - 1 CAAGUCCU--GCCUCCUUUUCGUCUCCUUGCCAGCCAUCUUCACCUUCUUGUCCCUGCCCAUGUCCGUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA ..((((((--((((((....((.....((((.(((((....(((...(.((........)).)...))).......))))).))))...)).))..))).)))))))............. ( -26.50) >consensus CAAGU____UACC______AGGUCCU_UUGCCUG__________GUCCUCUUCCCUGGCCAUGUCGCUGUCACACUUGGCUCGCAGUUUUGUGGCCAGGCCAGGGCUAUGAAAAUAACCA ............................................(((((....((((((((((..(((((..(....)....)))))..))))))))))..)))))(((....))).... (-24.19 = -24.88 + 0.69)


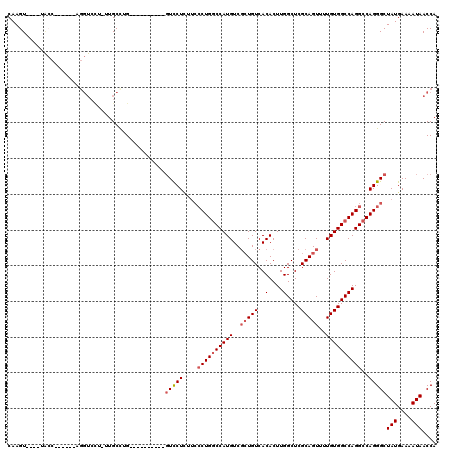
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:20 2006