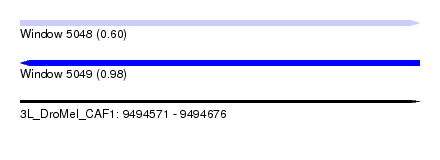
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,494,571 – 9,494,676 |
| Length | 105 |
| Max. P | 0.976084 |

| Location | 9,494,571 – 9,494,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -15.18 |
| Energy contribution | -14.15 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
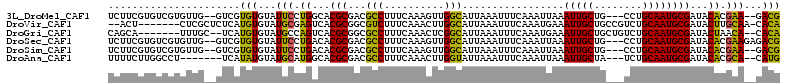
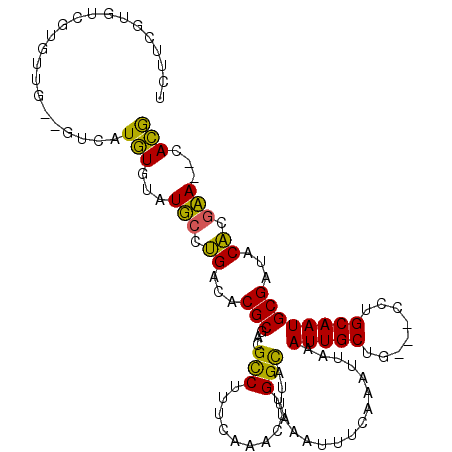
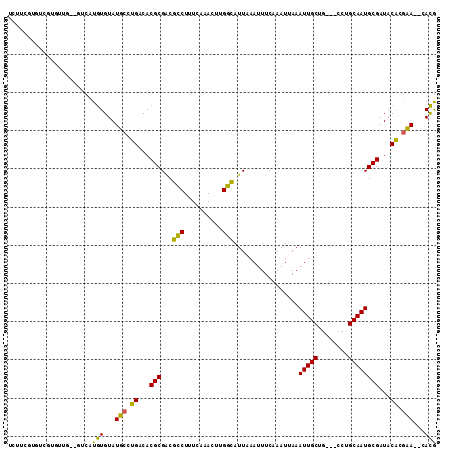
>3L_DroMel_CAF1 9494571 105 + 23771897 CGUC--UUCGUGUAUCGCAUUGCAGG---CAGCAAUUUAAUUUGAAAUUUAAUGCCAACUUUGAAAGGCGUCGCGUGCCAGGAAUACACACGAC--CAACACGACACGAAGA ..((--(((((((...((......((---((..(((((......)))))...))))..((.....))))((((.(((.........))).))))--.......))))))))) ( -28.10) >DroVir_CAF1 10611 102 + 1 UGUG-UUGCAAGUAUCGCAUUGCAGACGGCAGCAAUUUCAUUUGAAAUUUAAUGCCAAGUUUGAAAGACGCCGCGUGACUCGCAUACACAUGAGAGCGAG-------AGU-- ((((-((((.(((..(((...((....((((..((((((....))))))...))))..(((.....))))).)))..))).))).))))).....((...-------.))-- ( -28.40) >DroGri_CAF1 9127 101 + 1 UGUG--UGUUAGUAUCGCAUUGCAGACAGCAGCAAUUUCAUUUGAAAUUUAAUGCCGAGUUUGAAAGGCGCCGCGUGACUGGCAUACACAUGA--GCAAA-------UGCUG ((((--(((((((..(((..(((.....)))((((((((....))))))....(((..........))))).)))..)))))))))))....(--((...-------.))). ( -30.80) >DroSec_CAF1 11789 107 + 1 CGUCUCUUCGUGUAUCGCAUUGCAGG---CAGCAAUUUAAUUUGAAAUUUAAUGCCAACUUUGAAAGGCGUCGCGUGUCAGGAAUACACACGAC--CAACACGACACGAAGA ....(((((((((...((......((---((..(((((......)))))...))))..((.....))))((((.((((.......)))).))))--.......))))))))) ( -29.60) >DroSim_CAF1 12157 105 + 1 CGUC--UUCGUGUAUCGCAUUGCAGG---CAGCAAUUUAAUUUGAAAUUUAAUGCCAACUUUGAAAGGCGUCGCGUGUCAGGAAUACACACGAC--CAACACGACACGAAGA ..((--(((((((...((......((---((..(((((......)))))...))))..((.....))))((((.((((.......)))).))))--.......))))))))) ( -29.70) >DroAna_CAF1 8802 100 + 1 CAUG--UGCGUGUAUCGCAUUGCAGA---UAGCAAUUUAAUUUGAAAUUUAAUACCAAGUUUGAAAGGCGUCGCGUGCCAUGCAUACAUAUGA-------AGGCCAAGAAAA ((((--((..(((((.((((.((.((---(.((..(((((((((...........)))).)))))..))))))))))).)))))..)))))).-------............ ( -25.40) >consensus CGUC__UGCGUGUAUCGCAUUGCAGA___CAGCAAUUUAAUUUGAAAUUUAAUGCCAACUUUGAAAGGCGUCGCGUGACAGGAAUACACACGAC__CAACACGACACGAAGA .((...(((.((...((((((((........)))))................((((..........))))..)))...)).)))...))....................... (-15.18 = -14.15 + -1.03)

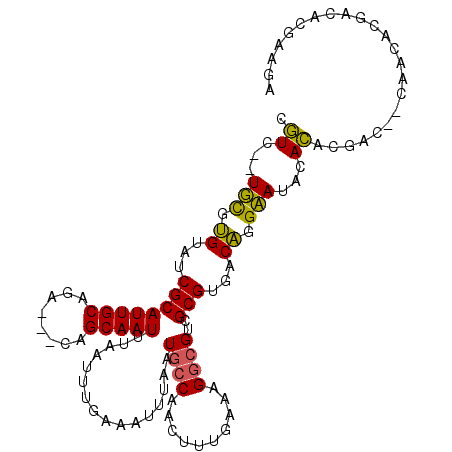
| Location | 9,494,571 – 9,494,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -15.78 |
| Energy contribution | -14.23 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9494571 105 - 23771897 UCUUCGUGUCGUGUUG--GUCGUGUGUAUUCCUGGCACGCGACGCCUUUCAAAGUUGGCAUUAAAUUUCAAAUUAAAUUGCUG---CCUGCAAUGCGAUACACGAA--GACG ((((((((((((((((--(((((((((.......))))))))).............((((...(((((......)))))..))---))..))))))))..))))))--)).. ( -39.40) >DroVir_CAF1 10611 102 - 1 --ACU-------CUCGCUCUCAUGUGUAUGCGAGUCACGCGGCGUCUUUCAAACUUGGCAUUAAAUUUCAAAUGAAAUUGCUGCCGUCUGCAAUGCGAUACUUGCAA-CACA --...-------..........(((((.(((((((..(((.((((.......))..((((...((((((....))))))..))))....))...)))..))))))))-)))) ( -28.60) >DroGri_CAF1 9127 101 - 1 CAGCA-------UUUGC--UCAUGUGUAUGCCAGUCACGCGGCGCCUUUCAAACUCGGCAUUAAAUUUCAAAUGAAAUUGCUGCUGUCUGCAAUGCGAUACUAACA--CACA ..(((-------(.(((--......)))(((.((.((.((((((((..........)))....((((((....))))))))))))).)))))))))..........--.... ( -25.70) >DroSec_CAF1 11789 107 - 1 UCUUCGUGUCGUGUUG--GUCGUGUGUAUUCCUGACACGCGACGCCUUUCAAAGUUGGCAUUAAAUUUCAAAUUAAAUUGCUG---CCUGCAAUGCGAUACACGAAGAGACG ((((((((((((((((--(((((((((.......))))))))).............((((...(((((......)))))..))---))..))))))))..)))))))).... ( -39.40) >DroSim_CAF1 12157 105 - 1 UCUUCGUGUCGUGUUG--GUCGUGUGUAUUCCUGACACGCGACGCCUUUCAAAGUUGGCAUUAAAUUUCAAAUUAAAUUGCUG---CCUGCAAUGCGAUACACGAA--GACG ((((((((((((((((--(((((((((.......))))))))).............((((...(((((......)))))..))---))..))))))))..))))))--)).. ( -39.10) >DroAna_CAF1 8802 100 - 1 UUUUCUUGGCCU-------UCAUAUGUAUGCAUGGCACGCGACGCCUUUCAAACUUGGUAUUAAAUUUCAAAUUAAAUUGCUA---UCUGCAAUGCGAUACACGCA--CAUG ........(((.-------.(((....)))...)))..(((..(((..........)))............(((..(((((..---...)))))..)))...))).--.... ( -16.60) >consensus UCUUCGUGUCGUGUUG__GUCAUGUGUAUGCCUGACACGCGACGCCUUUCAAACUUGGCAUUAAAUUUCAAAUUAAAUUGCUG___CCUGCAAUGCGAUACACGAA__CACG ......................(((...(((.((...(((...(((..........))).................(((((........))))))))...)).)))...))) (-15.78 = -14.23 + -1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:55 2006