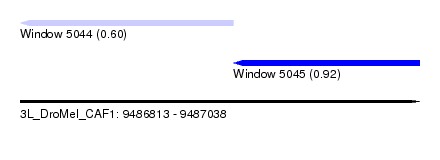
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,486,813 – 9,487,038 |
| Length | 225 |
| Max. P | 0.923250 |

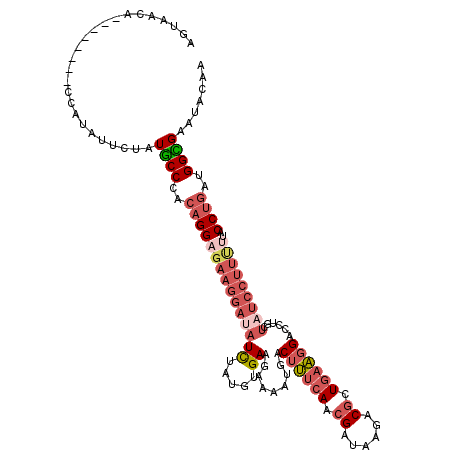
| Location | 9,486,813 – 9,486,933 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -16.77 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9486813 120 - 23771897 AGUAACACCAUAACACCAUAUUUUAUGCCCACAGGAGAAGGAUAUUUAUGUGAAGAAAAUGACUUUCAACGAUAAGACGCUGAAGGAUCUGUUAUCCUUUUUACCUGUUGGUGAAUACAA .(((.(((((......((((...))))...((((((((((((((.((((.........))))((((((.((......)).))))))......)))))))))..))))))))))..))).. ( -26.90) >DroSec_CAF1 4042 112 - 1 AGUAACA--------CCAUAUUCUAUGCCCACAGGAGAAGGAUAUCUAUGUGAAGAAAAUGACUUUCAACGAUAAGACGCUGAAGGACCUGCUAUCCUUUUUACCUGAUGGUGAAUACAA .(((.((--------((((............((((((((((((((((......)))......((((((.((......)).))))))......)))))))))..))))))))))..))).. ( -27.10) >DroSim_CAF1 3797 112 - 1 AGUAACA--------CCAUAUUCUAUGCCCACAGGAGAAGGAUAUCUAUGUGAAGAAAAUGACUUUCAACGAUAAGACGCUGAAGGACCUGUUAUCCUUUUUACCUGAUGGCGAAUACAA .......--------...(((((...(((..((((((((((((((((......)))......((((((.((......)).))))))......)))))))))..))))..))))))))... ( -27.30) >DroEre_CAF1 3610 111 - 1 AGUAUCG--------CC-UUUUGUAUGCCCACAGGAGAAGGAUAUCUAUGUGAAAAGAAUGACUUUCAACGAUAAGACGGUGAAGGACCUGCUUUCCUUCCUACCUGAUGGCGAAUACAA .((((((--------((-...((......))((((.((((((..(((........)))....((((((.((......)).))))))........))))))...))))..)))).)))).. ( -28.60) >DroYak_CAF1 5815 111 - 1 AGUAUCG--------CC-UUUUCUAUGCCAACAGGAGAAGGAUAUCUAUGUGAAGAAAAUGACUUUCAACGAUAAGACGGUGAAGGACCUGCUCUCCUUCUUACCUGAUGGCGAGUACAA .......--------..-.......(((((.(((((((((((..(((......)))......((((((.((......)).))))))........)))))))..)))).)))))....... ( -29.60) >DroPer_CAF1 9425 104 - 1 AUAAUCC--------CCAUCUU--UUUCCAACAGGAAAAGGAUAUCUUUCUGAAACAGAUGACGCUCAGUGAGAAGGCCGUCCGGGACCUGCCA------CUGCCCAAUGGGGAAUACAA ....(((--------(((((((--(((((....)))))))))).((..((((...)))).)).(..(((((.(.(((((.....)).))).)))------)))..)...)))))...... ( -34.40) >consensus AGUAACA________CCAUAUUCUAUGCCCACAGGAGAAGGAUAUCUAUGUGAAGAAAAUGACUUUCAACGAUAAGACGCUGAAGGACCUGCUAUCCUUUUUACCUGAUGGCGAAUACAA .........................((((..(((((((((((((((.....)).........((((((.((......)).))))))......)))))))))..))))..))))....... (-16.77 = -18.58 + 1.81)


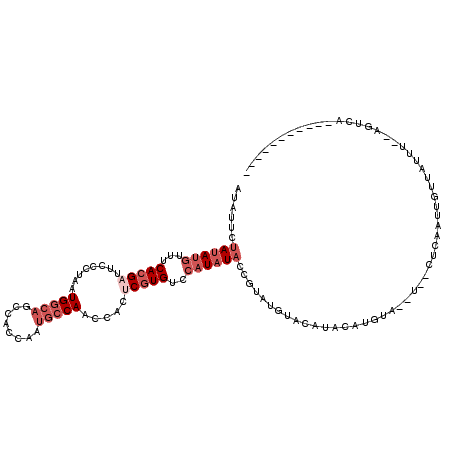
| Location | 9,486,933 – 9,487,038 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.06 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -8.75 |
| Energy contribution | -10.42 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9486933 105 - 23771897 AUAUUCUAUAUGUUUCACGAUUCCCUAAUGGCAGCCACCAAUGCCAACCACUCGUGUCCAUAUACAGUAAGUACAAACAUGUA--U--CUCAAUUGUUAUUUCACGUCA----------- .((((.((((((...(((((........(((((........))))).....)))))..)))))).)))).(((((....))))--)--.....................----------- ( -18.92) >DroSec_CAF1 4154 105 - 1 AUAUUCUAUAUGUUUCACGAUUCCCUAAUGGCAGCCACCAAUGCCAACCACUCGUGUCCAUAUACCGUAUGUACAAAGAUGUA--U--CUCAAUUGUUAUUUCAAUUCU----------- ((((..((((((...(((((........(((((........))))).....)))))..))))))..))))(((((....))))--)--...(((((......)))))..----------- ( -19.12) >DroSim_CAF1 3909 105 - 1 AUAUUCUAUAUGUUUCACGAUUCCCUAAUGGCAGCCACCAAUGCCAACCACUCGUGUCCAUAUACCGUAUGUACAUAGAUGUA--U--CUCAAUUGUUAUUUCAAGUCA----------- ((((..((((((...(((((........(((((........))))).....)))))..))))))..))))(((((....))))--)--.....................----------- ( -18.12) >DroEre_CAF1 3721 107 - 1 AUUUUCUAUAUGUUUCACGAUUCCUUAAUGGCAGCCACCAAUGCCAAUCACACGUGUCCAUAUACCGUAGGUACAUAUAUGUACAUAUCUCCAUUGCUUUUU--GGUUA----------- ......((((((...((((.........(((((........)))))......))))..))))))......(((((....)))))......(((........)--))...----------- ( -19.16) >DroYak_CAF1 5926 116 - 1 AUUUUCUAUAUGUUUCACGAUUCCUUAAUGGCAGCCACCAAUGCCAAUCACUCGUGUCCAUAUACCGUAUGUACAUAAAUAUA--UCUUUACACCAAAUAAC--AAUUAACAAUAACAAU ......((((((...(((((........(((((........))))).....)))))..))))))..((((((......)))))--)................--................ ( -16.42) >DroPer_CAF1 9529 100 - 1 CUGUUCUACAUAUUCCACGACUCUCUGAUGAGCGCCACAAAUGCCAAUCACACCUGUCCCUAUCAGGUAAGUCGUGGCUAUCC--U--UUCUCUCACUC-----AGUGU----------- ..............((((((((...((((..(((.......)))..)))).(((((.......))))).))))))))......--.--...........-----.....----------- ( -22.10) >consensus AUAUUCUAUAUGUUUCACGAUUCCCUAAUGGCAGCCACCAAUGCCAACCACUCGUGUCCAUAUACCGUAUGUACAUACAUGUA__U__CUCAAUUGUUAUUU__AGUCA___________ ......((((((...(((((........(((((........))))).....)))))..))))))........................................................ ( -8.75 = -10.42 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:52 2006