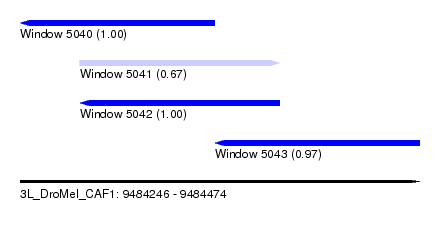
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,484,246 – 9,484,474 |
| Length | 228 |
| Max. P | 0.999926 |

| Location | 9,484,246 – 9,484,357 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -21.14 |
| Energy contribution | -20.14 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9484246 111 - 23771897 ACUCUGUUCUGUUCAGUAAGACCACUUGGGAGUCGAACAGG-CGCCAAUG--CUUGUACCGACUCUCAACGUCUGUAGAUUGCGCAUGUGACUCCG------UUCACACCAAUCGAAAAU ...........(((....((((...((((((((((.(((((-((....))--)))))..)))))))))).))))...(((((.(..(((((.....------.))))))))))))))... ( -34.60) >DroPse_CAF1 574 106 - 1 AAUG--UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG---CAAAUG--CCUGCACCGGCUCUCAACGUCUUUUGAUUGUGUAGAUGACUUUG------CUCACUCCAAUCUU-AAU ....--.((((.((((.(((((...((((((((((..((((---(....)--))))...)))))))))).))))))))).((.(((((....))))------).))...))))...-... ( -32.60) >DroAna_CAF1 1553 113 - 1 ACUCU-UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAAGUCGCUUAUGCGCUUGUACCGGCUCUCAACGUCUUUAGAUUGCUAUAGUGGUUCUA------GCCACUGCGAUCGUAAAU .....-...........(((((...((((((((((.(((((.(((....))))))))..)))))))))).)))))..((((((...((((((....------))))))))))))...... ( -42.60) >DroPer_CAF1 574 106 - 1 AAUG--UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG---CAAAUG--CCUGCACCGGCUCUCAACGUCUUUUGAUUGUGUAGAUGACUUUG------CUCACUCCAAUCUU-AAU ....--.((((.((((.(((((...((((((((((..((((---(....)--))))...)))))))))).))))))))).((.(((((....))))------).))...))))...-... ( -32.60) >DroWil_CAF1 1075 113 - 1 AAUUU-GAUUGUUCAGUAAGACCACUUGGGAGUCGAACAAG---CAUUGC--CUUGUACCGGCUUUCAACGUCUUUUGAUUAGGCAGUUGAUUUUGAAUAUAUUCACUCCAAUCAU-AAU ....(-((((..((((.(((((...(((..(((((.(((((---......--)))))..)))))..))).)))))))))...((.(((.((............)))))))))))).-... ( -27.60) >consensus AAUCU_UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG___CAAAUG__CUUGUACCGGCUCUCAACGUCUUUUGAUUGCGCAGAUGACUUUG______CUCACUCCAAUCGU_AAU .................(((((...((((((((((..(((((.........)))))...)))))))))).)))))..(((((.(....(((............)))..))))))...... (-21.14 = -20.14 + -1.00)

| Location | 9,484,280 – 9,484,394 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.44 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9484280 114 + 23771897 AUCUACAGACGUUGAGAGUCGGUACAAG--CAUUGGCG-CCUGUUCGACUCCCAAGUGGUCUUACUGAACAGAACAGAGUUUAUAUCUGAG--AGAGCCUGGCGACGA-UGUUGUCGCCU ........((.(((.(((((((.(((.(--(......)-).)))))))))).)))))((((((.((....))..((((.......)))).)--))).)).(((((((.-...))))))). ( -35.30) >DroPse_CAF1 607 112 + 1 AUCAAAAGACGUUGAGAGCCGGUGCAGG--CAUUUG---CCUGUUCGACUCCCAAGUGGUCUUACUGAACAACA--CAUUUUUCUUGUGAUUUAUUGCCUAGCGACGUUUGACGUCG-CU .(((.(((((.(((.(((.(((.(((((--(....)---)))))))).))).)))...)))))..)))....((--((.......))))...........(((((((.....)))))-)) ( -36.70) >DroAna_CAF1 1587 114 + 1 AUCUAAAGACGUUGAGAGCCGGUACAAGCGCAUAAGCGACUUGUUCGACUCCCAAGUGGUCUUACUGAACAACA-AGAGUUAUCUUCUCU----ACACAUGGCGACUC-UCUGGUCGCCU .((..(((((.(((.(((.(((.((((((((....))).)))))))).))).)))...)))))...))......-((((.......))))----......(((((((.-...))))))). ( -35.80) >DroPer_CAF1 607 112 + 1 AUCAAAAGACGUUGAGAGCCGGUGCAGG--CAUUUG---CCUGUUCGACUCCCAAGUGGUCUUACUGAACAACA--CAUUUUUCUUGUGAUUUAUUGCCUAGCGACGUUUGACGUCG-CU .(((.(((((.(((.(((.(((.(((((--(....)---)))))))).))).)))...)))))..)))....((--((.......))))...........(((((((.....)))))-)) ( -36.70) >DroWil_CAF1 1114 98 + 1 AUCAAAAGACGUUGAAAGCCGGUACAAG--GCAAUG---CUUGUUCGACUCCCAAGUGGUCUUACUGAACAAUC-AAAUUAAU--UCUC----AU----UGGCAA-----AACGCCA-UU .(((.(((((.(((..((.(((.(((((--......---)))))))).))..)))...)))))..)))......-........--....----..----((((..-----...))))-.. ( -18.60) >consensus AUCAAAAGACGUUGAGAGCCGGUACAAG__CAUUUG___CCUGUUCGACUCCCAAGUGGUCUUACUGAACAACA_ACAUUUUUCUUCUGA___AUUGCCUGGCGACGU_UGACGUCG_CU .....(((((.(((.(((.(((.(((((...........)))))))).))).)))...))))).....................................(((((((.....))))).)) (-15.52 = -15.84 + 0.32)

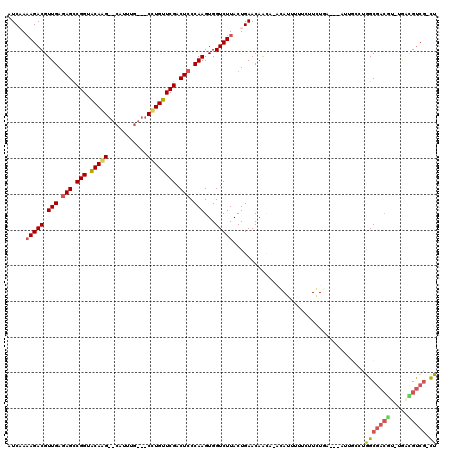
| Location | 9,484,280 – 9,484,394 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.44 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -22.20 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.56 |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9484280 114 - 23771897 AGGCGACAACA-UCGUCGCCAGGCUCU--CUCAGAUAUAAACUCUGUUCUGUUCAGUAAGACCACUUGGGAGUCGAACAGG-CGCCAAUG--CUUGUACCGACUCUCAACGUCUGUAGAU .((((((....-..))))))......(--(((((((........((.(((........))).)).((((((((((.(((((-((....))--)))))..)))))))))).))))).))). ( -41.80) >DroPse_CAF1 607 112 - 1 AG-CGACGUCAAACGUCGCUAGGCAAUAAAUCACAAGAAAAAUG--UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG---CAAAUG--CCUGCACCGGCUCUCAACGUCUUUUGAU ((-(((((.....)))))))..((((.....((((.......))--))))))((((.(((((...((((((((((..((((---(....)--))))...)))))))))).))))))))). ( -42.60) >DroAna_CAF1 1587 114 - 1 AGGCGACCAGA-GAGUCGCCAUGUGU----AGAGAAGAUAACUCU-UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAAGUCGCUUAUGCGCUUGUACCGGCUCUCAACGUCUUUAGAU .((((((....-..))))))..((.(----(((((.((((((...-.))))))............((((((((((.(((((.(((....))))))))..))))))))))..)))))).)) ( -43.10) >DroPer_CAF1 607 112 - 1 AG-CGACGUCAAACGUCGCUAGGCAAUAAAUCACAAGAAAAAUG--UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG---CAAAUG--CCUGCACCGGCUCUCAACGUCUUUUGAU ((-(((((.....)))))))..((((.....((((.......))--))))))((((.(((((...((((((((((..((((---(....)--))))...)))))))))).))))))))). ( -42.60) >DroWil_CAF1 1114 98 - 1 AA-UGGCGUU-----UUGCCA----AU----GAGA--AUUAAUUU-GAUUGUUCAGUAAGACCACUUGGGAGUCGAACAAG---CAUUGC--CUUGUACCGGCUUUCAACGUCUUUUGAU ..-((((...-----..))))----..----....--........-......((((.(((((...(((..(((((.(((((---......--)))))..)))))..))).))))))))). ( -26.60) >consensus AG_CGACGUCA_ACGUCGCCAGGCAAU___UCAGAAGAAAAAUCU_UGUUGUUCAGUAAGACCACUUGGGAGUCGAACAGG___CAAAUG__CUUGUACCGGCUCUCAACGUCUUUUGAU ...((((.......))))..................................((((.(((((...((((((((((..(((((.........)))))...)))))))))).))))))))). (-22.20 = -21.92 + -0.28)


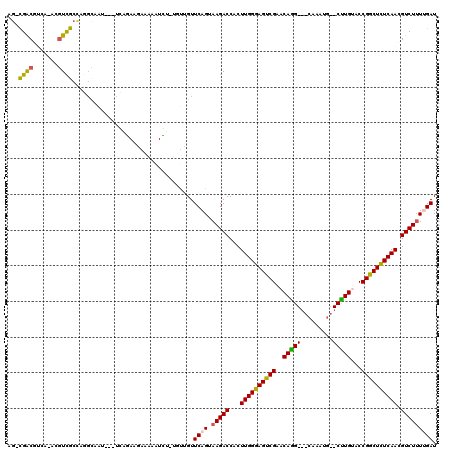
| Location | 9,484,357 – 9,484,474 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9484357 117 - 23771897 CAAGCUGGGACUUGCCAAGUCCAUCCAUCAUGCUCGCGUCCUGAUCCGUCAGCGUCACAUUCGGUAAAUAUCGCUUGGACAGGCGACAACA-UCGUCGCCAGGCUCU--CUCAGAUAUAA ....((((((...(((..(((((..(...(((((.(((........))).))))).......(((....))))..))))).((((((....-..)))))).)))..)--)))))...... ( -40.00) >DroPse_CAF1 680 119 - 1 CAAGCUUGGAUUGGCCAAGUCCAUCCAUCAUGCCCGUGUGCUGAUCCGCCAGCGUCACAUUCGGUAAAUAUCGCUUGGACAG-CGACGUCAAACGUCGCUAGGCAAUAAAUCACAAGAAA ....((((((((.(((..(((((.......((((.((((((((......))))).)))....)))).........)))))((-(((((.....))))))).)))....)))).))))... ( -42.99) >DroAna_CAF1 1666 115 - 1 CAAGCUUGGACUUGCCAAGUCCAUCCACCACGCCCGUGUGCUGAUCCGCCAGCGUCACAUUCGGUAAAUAUCGCUUGGACAGGCGACCAGA-GAGUCGCCAUGUGU----AGAGAAGAUA (((((.((((((.....))))))........(((.((((((((......))))).)))....))).......))))).(((((((((....-..)))))).)))..----.......... ( -37.40) >DroPer_CAF1 680 119 - 1 CAAGCUUGGAUUGGCCAAGUCCAUCCAUCAUGCCCGUGUGCUGAUCCGCCAGCGUCACAUUCGGUAAAUAUCGCUUGGACAG-CGACGUCAAACGUCGCUAGGCAAUAAAUCACAAGAAA ....((((((((.(((..(((((.......((((.((((((((......))))).)))....)))).........)))))((-(((((.....))))))).)))....)))).))))... ( -42.99) >DroWil_CAF1 1188 104 - 1 CAAGCUGGGUUUGGCUAAAUCCAUCCAUCAUGCCCGUGUGCUGAUCCGUCAGCGUCACAUUCGGUAAAUAUCGCUGGGCCAA-UGGCGUU-----UUGCCA----AU----GAGA--AUU .....(((((((....)))))))....(((((((((((((((((....)))))).)))...((((....))))..))))...-((((...-----..))))----))----))..--... ( -31.90) >consensus CAAGCUUGGAUUGGCCAAGUCCAUCCAUCAUGCCCGUGUGCUGAUCCGCCAGCGUCACAUUCGGUAAAUAUCGCUUGGACAG_CGACGUCA_ACGUCGCCAGGCAAU___UCAGAAGAAA (((((..(((((.....)))))........((((.((((((((......))))).)))....))))......)))))......((((.......))))...................... (-22.18 = -22.34 + 0.16)

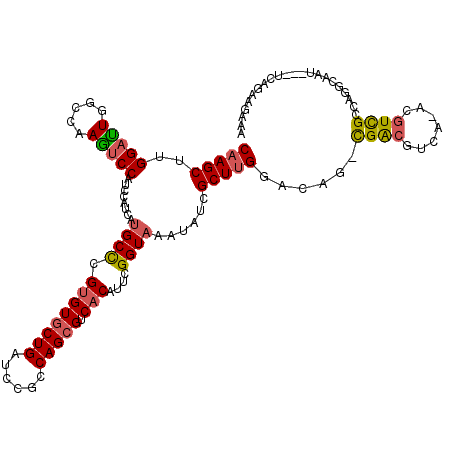

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:50 2006