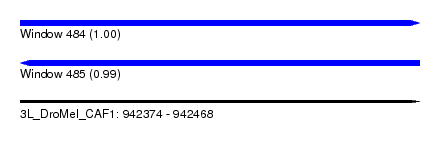
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 942,374 – 942,468 |
| Length | 94 |
| Max. P | 0.998844 |

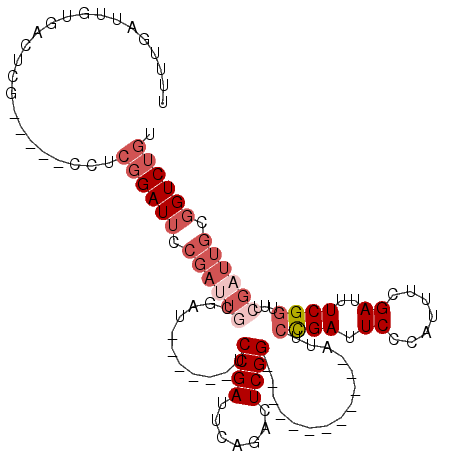
| Location | 942,374 – 942,468 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -6.58 |
| Energy contribution | -6.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.24 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 942374 94 + 23771897 ACAGACCGCAAUCGAAACCGAAAUCGAAAUGGGAAUCAGAAU------------CCGAGUCUGAAUCGGA------AUCGGAAUCGGAAUCCGAGG-----CGAGUCACAAUCAAAA ...((((((..((((........))))....(((.((.((.(------------((((.((((...))))------.))))).)).)).)))...)-----)).))).......... ( -28.80) >DroSec_CAF1 77420 82 + 1 ACAGAC------------CGAAAUCGAAGUGGGAAUCGGAAU------------CCGAGUCUGAAUCGGA------AUCUGAAUCGGAAUCCGAGG-----CGACUCACAAUCAAAA .....(------------(((..((......))..))))...------------..(((((.(..(((((------.((((...)))).)))))..-----)))))).......... ( -23.80) >DroSim_CAF1 69330 100 + 1 ACAGACCGCAAUCGAAACCGAAAUCGAAAUGGGAAUCGGAAU------------CCGAGUCUGAAUCGGAAUCGGAAUCUCAAUCGGAAUCCGAGG-----CGAGUCACAAUCAAAA ...((((((..((((........))))..(((((.((.((.(------------((((.......))))).)).)).))))).((((...)))).)-----)).))).......... ( -31.20) >DroEre_CAF1 79320 111 + 1 ACAGACCGCAAUCGAAACCGAAAUCGAAAUGCGAAUCGGCAUCCCAAUCCGCAUCCGAAUGUGAAUCGGA------UUCGCAAUCGCAAUCCGAGGCAAUGCGAGUCACAAUCAAAA ...(((((((.(((....)))..(((..(((((.((.((....)).)).))))).)))...((..(((((------((.((....)))))))))..)).)))).))).......... ( -32.60) >DroYak_CAF1 81247 83 + 1 ACAGACCGCAAUCGAAACCGAAAUCGAAAUGGGAAUCGGAAU------------CCGAAUCUGAAUCGGA------AUCGCA-UCGCAAUC----GCAAUCCGAGG----------- .....(((...((((........))))..)))...(((((.(------------((((.......)))))------......-..((....----))..)))))..----------- ( -18.60) >consensus ACAGACCGCAAUCGAAACCGAAAUCGAAAUGGGAAUCGGAAU____________CCGAGUCUGAAUCGGA______AUCGCAAUCGGAAUCCGAGG_____CGAGUCACAAUCAAAA ..................(((..((((........))))...............((((.......))))..............)))............................... ( -6.58 = -6.78 + 0.20)

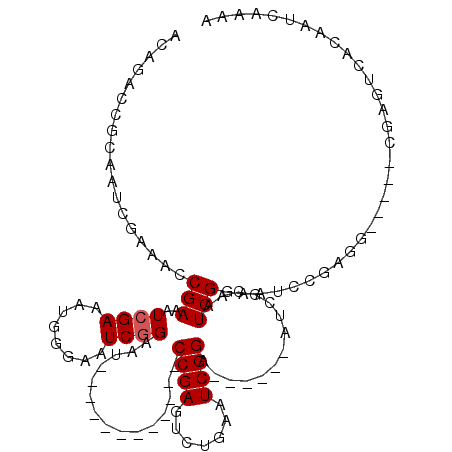
| Location | 942,374 – 942,468 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.12 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -9.62 |
| Energy contribution | -11.86 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 942374 94 - 23771897 UUUUGAUUGUGACUCG-----CCUCGGAUUCCGAUUCCGAU------UCCGAUUCAGACUCGG------------AUUCUGAUUCCCAUUUCGAUUUCGGUUUCGAUUGCGGUCUGU ..........(((.((-----(.(((((..((((...(((.------...((.(((((.....------------..))))).)).....)))...)))).)))))..))))))... ( -26.20) >DroSec_CAF1 77420 82 - 1 UUUUGAUUGUGAGUCG-----CCUCGGAUUCCGAUUCAGAU------UCCGAUUCAGACUCGG------------AUUCCGAUUCCCACUUCGAUUUCG------------GUCUGU ....(((((.(((((.-----..((((...))))....)))------)))))))(((((.(((------------(...(((........)))..))))------------))))). ( -23.50) >DroSim_CAF1 69330 100 - 1 UUUUGAUUGUGACUCG-----CCUCGGAUUCCGAUUGAGAUUCCGAUUCCGAUUCAGACUCGG------------AUUCCGAUUCCCAUUUCGAUUUCGGUUUCGAUUGCGGUCUGU ..........(((.((-----(.(((((..(((((((((((((.((.(((((.......))))------------).)).)).....)))))))..)))).)))))..))))))... ( -29.20) >DroEre_CAF1 79320 111 - 1 UUUUGAUUGUGACUCGCAUUGCCUCGGAUUGCGAUUGCGAA------UCCGAUUCACAUUCGGAUGCGGAUUGGGAUGCCGAUUCGCAUUUCGAUUUCGGUUUCGAUUGCGGUCUGU ....(((((..(.(((....((((((((((((....)).))------))))).......(((((((((((((((....)))))))))).)))))....)))..))))..)))))... ( -41.30) >DroYak_CAF1 81247 83 - 1 -----------CCUCGGAUUGC----GAUUGCGA-UGCGAU------UCCGAUUCAGAUUCGG------------AUUCCGAUUCCCAUUUCGAUUUCGGUUUCGAUUGCGGUCUGU -----------...((((((((----(((((.((-(.(((.------..(((....((.((((------------...)))).)).....)))...)))))).))))))))))))). ( -27.40) >consensus UUUUGAUUGUGACUCG_____CCUCGGAUUCCGAUUGCGAU______UCCGAUUCAGACUCGG____________AUUCCGAUUCCCAUUUCGAUUUCGGUUUCGAUUGCGGUCUGU ........................((((((.((((((...........((((.......))))...............((((.((.......))..))))...)))))).)))))). ( -9.62 = -11.86 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:15 2006