
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,474,628 – 9,474,758 |
| Length | 130 |
| Max. P | 0.979426 |

| Location | 9,474,628 – 9,474,727 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -11.75 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
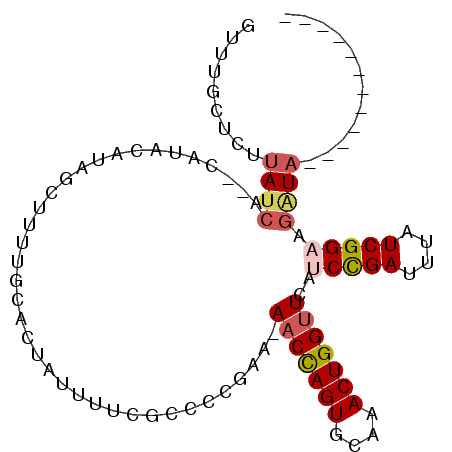
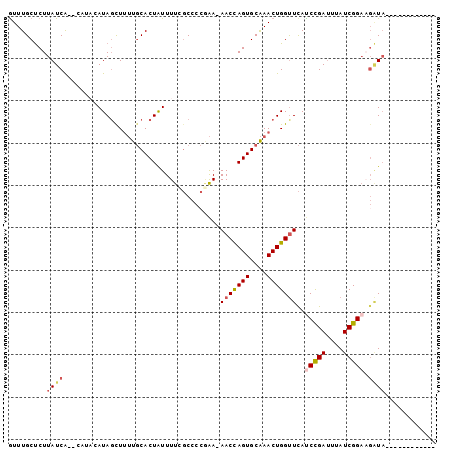
>3L_DroMel_CAF1 9474628 99 + 23771897 AUUUGCUCUUAUCAUACAUACAUACCUUUUGCACUAUUUUCGCCCCGAAAAACCAGUGCAAACUGGUUCAUCCGAUUUAUCGGAAGAUAUAC--UACACCU .........((((..........(((.((((((((.((((((...))))))...))))))))..)))...(((((....))))).))))...--....... ( -22.80) >DroSec_CAF1 12305 98 + 1 GUUUGCUCUUAUCA--CAUACAUAGCUUUUGCACUAUUUUCGCCCCGAA-AACCAGUGCAAACUGGUUCAUCCGAUAGAUCGGAAGAUAUACCCCACACCU ((.((....((((.--.......((((((((((((.((((((...))))-))..))))))))..))))..(((((....))))).)))).....)).)).. ( -21.90) >DroSim_CAF1 12911 98 + 1 GUUUGCUCUUAUCA--CAUACAUAGCUUUUGCACUAUUUUCGCCCCGAA-AACCAGUGCAAACUGGUUCAUCCGAUAGAUCGGAAGAUAUACCCCACACCU ((.((....((((.--.......((((((((((((.((((((...))))-))..))))))))..))))..(((((....))))).)))).....)).)).. ( -21.90) >DroEre_CAF1 12334 82 + 1 GUUUGCUCUUAUCG--C----AUAGUUUUUGCACUAUUUUAGCCCCAAA-AACCAGUGCAAACUGGUUCACCCGAUUUAUCGGAAGAUA------------ .........(((((--(----(.......))).................-(((((((....)))))))...((((....))))..))))------------ ( -16.80) >DroYak_CAF1 12432 85 + 1 GUUUGCUCUUAUCG--CACACAUAGUUUUUGCACUAUUUUUGCCCCAAA-AACCAGUGCAAACUGGCUCAUCCGAUUUAUCGGAU-CUA------------ ((.(((.......)--)).)).(((((..((((((.((((((...))))-))..)))))))))))....((((((....))))))-...------------ ( -21.10) >DroAna_CAF1 12741 77 + 1 GUUUGCCCUUAUCA--C----AUAGUU--UUCACUGUUUUUCCCC--GA-AACUAGUGCGAACUGGUUUAUCUGAACAAUCGGUCGGU------------- ....(((...(((.--.----.(((((--(.(((((((((.....--))-))).)))).))))))((((....))))....))).)))------------- ( -14.90) >consensus GUUUGCUCUUAUCA__CAUACAUAGCUUUUGCACUAUUUUCGCCCCGAA_AACCAGUGCAAACUGGUUCAUCCGAUUUAUCGGAAGAUA____________ .........((((.....................................(((((((....)))))))..(((((....))))).))))............ (-11.75 = -12.37 + 0.61)

| Location | 9,474,667 – 9,474,758 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 71.22 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
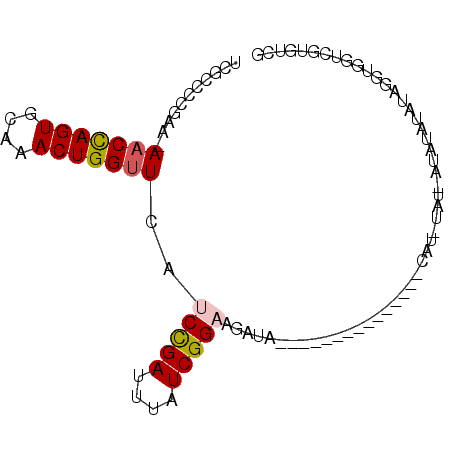
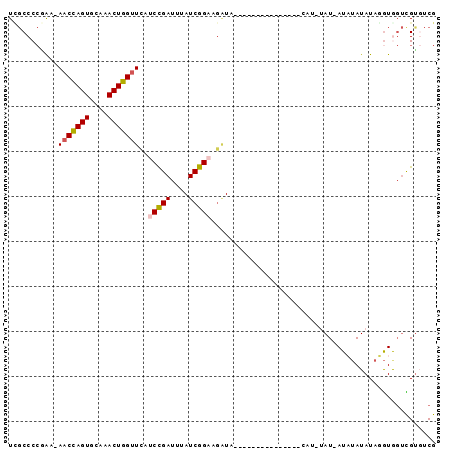
>3L_DroMel_CAF1 9474667 91 + 23771897 UCGCCCCGAAAAACCAGUGCAAACUGGUUCAUCCGAUUUAUCGGAAGAUAUAC--UACACCUAUUCAUAUAU-AUACAUAUAUAUAGUUGC-UCG ..((...(((..((((((....))))))((.(((((....))))).)).....--........))).(((((-((....)))))))...))-... ( -19.50) >DroSec_CAF1 12342 93 + 1 UCGCCCCGAA-AACCAGUGCAAACUGGUUCAUCCGAUAGAUCGGAAGAUAUACCCCACACCUAUUCAUAUAU-AUCUAUAUAGGUGGUCGUGUCG ..........-(((((((....)))))))..(((((....))))).(((((..((...((((((........-......))))))))..))))). ( -25.04) >DroSim_CAF1 12948 93 + 1 UCGCCCCGAA-AACCAGUGCAAACUGGUUCAUCCGAUAGAUCGGAAGAUAUACCCCACACCUAUUCAUAUAU-AUCUAUAUAGGUGGUCGUGUCG ..........-(((((((....)))))))..(((((....))))).(((((..((...((((((........-......))))))))..))))). ( -25.04) >DroEre_CAF1 12367 74 + 1 UAGCCCCAAA-AACCAGUGCAAACUGGUUCACCCGAUUUAUCGGAAGAUA-------------------UAU-AUAUAUAUAUGUGGUCGUGUCG ..........-(((((((....)))))))(((((((....))))..((((-------------------(((-((....)))))).))))))... ( -17.90) >DroYak_CAF1 12469 74 + 1 UUGCCCCAAA-AACCAGUGCAAACUGGCUCAUCCGAUUUAUCGGAU-CUA-------------------UAUCAUAUAUAUAGGUAGUCGUGUCG (((((.....-..(((((....)))))...((((((....))))))-...-------------------.............)))))........ ( -16.50) >DroAna_CAF1 12772 73 + 1 UUCCCC--GA-AACUAGUGCGAACUGGUUUAUCUGAACAAUCGGUCGGU----------------CGG--AU-AUAUAUAUAUGUGCAUGUCCCG ....((--((-(((((((....)))))))...((((....)))))))).----------------.((--((-((.(((....))).)))))).. ( -18.30) >consensus UCGCCCCGAA_AACCAGUGCAAACUGGUUCAUCCGAUUUAUCGGAAGAUA_______________CAU_UAU_AUAUAUAUAGGUGGUCGUGUCG ...........(((((((....)))))))..(((((....))))).................................................. (-11.36 = -11.58 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:42 2006