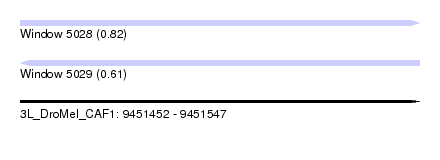
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,451,452 – 9,451,547 |
| Length | 95 |
| Max. P | 0.821438 |

| Location | 9,451,452 – 9,451,547 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
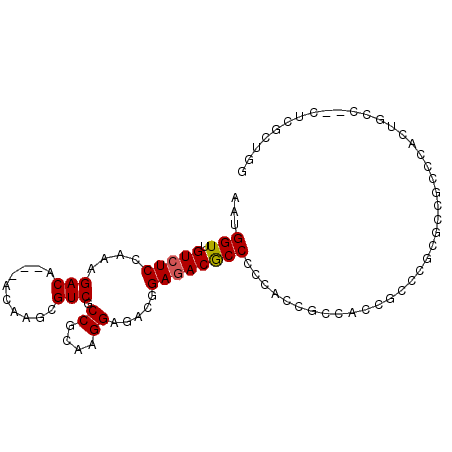
>3L_DroMel_CAF1 9451452 95 + 23771897 AAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACAGAGACGCCCCCACCGCCUCCGCCCGUACCGCCCGCUGGCUCCUCGCUGG ....(((((((....)))))))....((((((.((....)).))).(((..((.......))....(((.((.......)).)))..)))))).. ( -28.60) >DroVir_CAF1 11343 83 + 1 AAUGGUUGUCUCAAAAGACA---ACAAGCGUCGCCGUAAGGAAACGGAGACACCGCCAACGUCAUCGUCGGUGGCGGC---------GGCGGUGA ....(((((((....)))))---))...(((..((....))..)))....(((((((..((((((.....))))))..---------))))))). ( -40.10) >DroGri_CAF1 4556 83 + 1 AAUGGUUGUCUCAAAAGACA---ACAAGCGUCGCCGUAAGGAAACAGAUACGCCGCCAACGUCAUCGUCCGCGGCGUC---------GGCGGCGG ....(((((((....)))))---))...((((((((...(....)....(((((((..(((....)))..))))))))---------))))))). ( -41.00) >DroSec_CAF1 5789 95 + 1 AAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCGCGCCGCCCACUGCCUCCUCGCUGG ....(((((((....))))))).....(((((.((....)).)))((((.((.......)).))))))(((((..((.....)).....))).)) ( -29.10) >DroSim_CAF1 4102 95 + 1 AAUGGUUGUCUCCAAAGACAACAACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCUCCGCCCGCCCCGCCCACUGCCUCCUCGCUGG ....(((((((....)))))))....((((((.((....)).)))((((.((.......)).))))........................))).. ( -27.80) >DroAna_CAF1 4903 87 + 1 AAUGGUUGUCUCCAAAGACA---ACAAGCGUCGCCGCAAGGAGACGGAGACACCUCCCCCUCCACCACCAACGGCGUCCAAUGCU-----GCAGC ....(((((((....)))))---)).((((((((((...((((..((((....))))..))))........)))))....)))))-----..... ( -28.90) >consensus AAUGGUUGUCUCCAAAGACA___ACAAGCGUCGCCGCAAGGAGACGGAGACGCCCCCACCGCCACCGCCCGCGCCGCCCACUGCC__CUCGCUGG ...(((.(((((....(((..........))).((....)).....))))))))......................................... (-14.37 = -14.98 + 0.61)

| Location | 9,451,452 – 9,451,547 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
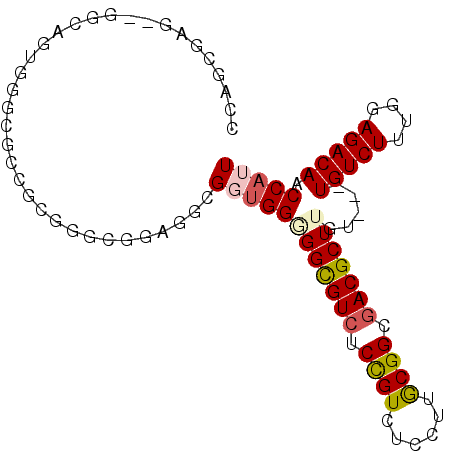
>3L_DroMel_CAF1 9451452 95 - 23771897 CCAGCGAGGAGCCAGCGGGCGGUACGGGCGGAGGCGGUGGGGGCGUCUCUGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUU ...((((((((((....)))......((((((((((.......)))))))))))))))))(((((((.....))))))).(((....)))..... ( -40.50) >DroVir_CAF1 11343 83 - 1 UCACCGCC---------GCCGCCACCGACGAUGACGUUGGCGGUGUCUCCGUUUCCUUACGGCGACGCUUGU---UGUCUUUUGAGACAACCAUU .....((.---------(((((((.((.......)).)))))))(((.((((......)))).)))))..((---(((((....))))))).... ( -32.60) >DroGri_CAF1 4556 83 - 1 CCGCCGCC---------GACGCCGCGGACGAUGACGUUGGCGGCGUAUCUGUUUCCUUACGGCGACGCUUGU---UGUCUUUUGAGACAACCAUU .((.((((---------.(((((((.((((....)))).)))))))....((......)))))).))...((---(((((....))))))).... ( -36.20) >DroSec_CAF1 5789 95 - 1 CCAGCGAGGAGGCAGUGGGCGGCGCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUU ...((((((((((.(((.....)))....(((((((.......)))))))))))))))))(((((((.....))))))).(((....)))..... ( -42.00) >DroSim_CAF1 4102 95 - 1 CCAGCGAGGAGGCAGUGGGCGGGGCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGUUGUUGUCUUUGGAGACAACCAUU ...((......))(((((((((((.(((((((((((.......)))))))))))))))))(((((((.....))))))).(((....)))))))) ( -43.20) >DroAna_CAF1 4903 87 - 1 GCUGC-----AGCAUUGGACGCCGUUGGUGGUGGAGGGGGAGGUGUCUCCGUCUCCUUGCGGCGACGCUUGU---UGUCUUUGGAGACAACCAUU (((((-----((.......((((...))))..(((((.((((....)))).)))))))))))).......((---(((((....))))))).... ( -36.00) >consensus CCAGCGAG__GGCAGUGGGCGCCGCGGGCGGAGGCGGUGGGGGCGUCUCCGUCUCCUUGCGGCGACGCUUGU___UGUCUUUGGAGACAACCAUU ...................................((((((((((((.((((......)))).))))))).....(((((....))))).))))) (-20.70 = -20.95 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:36 2006