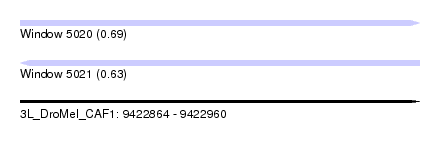
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,422,864 – 9,422,960 |
| Length | 96 |
| Max. P | 0.686736 |

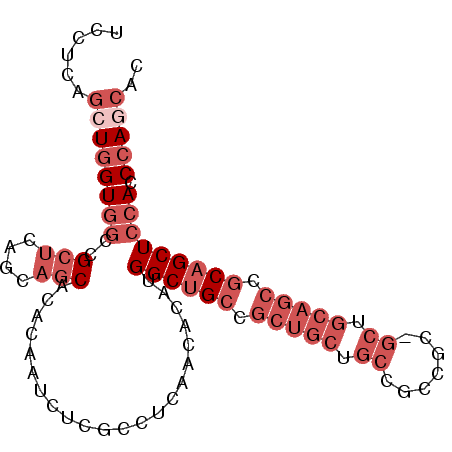
| Location | 9,422,864 – 9,422,960 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -32.51 |
| Consensus MFE | -21.73 |
| Energy contribution | -24.65 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9422864 96 + 23771897 CCCUCAGCUGGUGGCCGCUCAGCAGCACACAAUCUCGCCUCAACACAUGGCUGCCGCUGCUGCCGCCGCAGCUGCAGCCGCAGCUCCACCCAGCAC ......((((((((..(((..((.((..........))..........((((((.(((((.......))))).))))))))))).))).))))).. ( -38.90) >DroSec_CAF1 1928 84 + 1 UCCUCAGCUGGUGACCGCUCAGCAGCACACAAUCUCGCCUCAACACAUGGCUGCCGCUGCU------------GCAGCCGCAGCUCCACCCAGCAC ......(((((((...(((....)))......................((((((.((((..------------.)))).)))))).)).))))).. ( -26.50) >DroSim_CAF1 531 96 + 1 UCCUCAGCUGGUGGCCGCUCAGCAGCACACAAUCUCGCCUCAACACAUGGCUGCCGCUGCUGCCGCCGCAGCUGCAGCCGCAGCUCCACCCAGCAC ......((((((((..(((..((.((..........))..........((((((.(((((.......))))).))))))))))).))).))))).. ( -38.90) >DroEre_CAF1 1925 96 + 1 UCCUCAGAUGGUGGCCGCUCAGCAGCACACAAUCUCGCCCCAACACAUGGCUGCCGCUGCCGCCGCCGCUGCUGCAGCAUCAGCUCCAGCCAGCAC .........((((((.((.(.(((((.((..................))))))).)..)).))))))(((((((.(((....))).))).)))).. ( -35.97) >DroYak_CAF1 2232 96 + 1 UCCUCAGAUGGUGGCCGCUCAGCAGCACACAAUCUCGCCUCAACACAUGGCUGCCGCUGCCGCCGCCGCUGCUGCAGCAGCAGCUCCACCCAGCAC .........((((((.((..(((.(((.........(((.........)))))).))))).))))))(((((....))))).(((......))).. ( -34.10) >DroAna_CAF1 1979 81 + 1 UCCCCAACUGGUAGCCGCCCAGCAACACACAAUAUCGCCCCAGCACAUGGCUGCAGCCGCUGCC---------------GCUGCUCCAACAAGCUC ........(((.(((.((.((((.............((....))....(((....)))))))..---------------)).))))))........ ( -20.70) >consensus UCCUCAGCUGGUGGCCGCUCAGCAGCACACAAUCUCGCCUCAACACAUGGCUGCCGCUGCUGCCGCCGC_GCUGCAGCCGCAGCUCCACCCAGCAC ......((((((((..(((....)))......................((((((.(((((.((.......)).))))).))))))))).))))).. (-21.73 = -24.65 + 2.92)

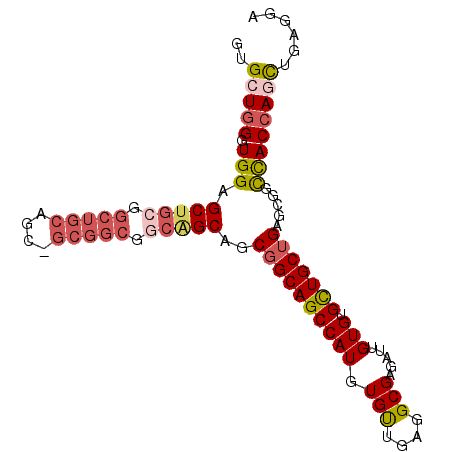
| Location | 9,422,864 – 9,422,960 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -32.57 |
| Energy contribution | -34.08 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9422864 96 - 23771897 GUGCUGGGUGGAGCUGCGGCUGCAGCUGCGGCGGCAGCAGCGGCAGCCAUGUGUUGAGGCGAGAUUGUGUGCUGCUGAGCGGCCACCAGCUGAGGG ..(((((.(((.(((((.(((((....))))).)))))..((((((((((((.(((...))).).)))).))))))).....))))))))...... ( -46.90) >DroSec_CAF1 1928 84 - 1 GUGCUGGGUGGAGCUGCGGCUGC------------AGCAGCGGCAGCCAUGUGUUGAGGCGAGAUUGUGUGCUGCUGAGCGGUCACCAGCUGAGGA ..(((((((......))((((((------------..((((((((..((..(((....)))....))..)))))))).)))))).)))))...... ( -33.80) >DroSim_CAF1 531 96 - 1 GUGCUGGGUGGAGCUGCGGCUGCAGCUGCGGCGGCAGCAGCGGCAGCCAUGUGUUGAGGCGAGAUUGUGUGCUGCUGAGCGGCCACCAGCUGAGGA ..(((((.(((.(((((.(((((....))))).)))))..((((((((((((.(((...))).).)))).))))))).....))))))))...... ( -46.90) >DroEre_CAF1 1925 96 - 1 GUGCUGGCUGGAGCUGAUGCUGCAGCAGCGGCGGCGGCAGCGGCAGCCAUGUGUUGGGGCGAGAUUGUGUGCUGCUGAGCGGCCACCAUCUGAGGA ..((((.(((.(((....))).)))))))(((.((..((((((((..((..(((....)))....))..)))))))).)).))).((......)). ( -44.30) >DroYak_CAF1 2232 96 - 1 GUGCUGGGUGGAGCUGCUGCUGCAGCAGCGGCGGCGGCAGCGGCAGCCAUGUGUUGAGGCGAGAUUGUGUGCUGCUGAGCGGCCACCAUCUGAGGA ...((((((((.((((((((....))))))))(((.((..((((((((((((.(((...))).).)))).))))))).)).))).)))))).)).. ( -47.70) >DroAna_CAF1 1979 81 - 1 GAGCUUGUUGGAGCAGC---------------GGCAGCGGCUGCAGCCAUGUGCUGGGGCGAUAUUGUGUGUUGCUGGGCGGCUACCAGUUGGGGA ...((..(((((((.((---------------..((((.(((.((((.....)))).)))(((((...))))))))).)).))).))))..))... ( -28.30) >consensus GUGCUGGGUGGAGCUGCGGCUGCAGC_GCGGCGGCAGCAGCGGCAGCCAUGUGUUGAGGCGAGAUUGUGUGCUGCUGAGCGGCCACCAGCUGAGGA ..(((((.(((.(((((.(((((....))))).)))))..((((((((((.(((....))).....))).))))))).....))))))))...... (-32.57 = -34.08 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:28 2006