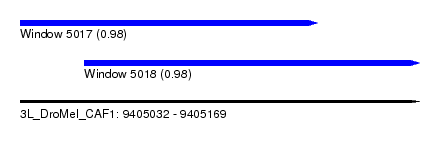
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,405,032 – 9,405,169 |
| Length | 137 |
| Max. P | 0.984509 |

| Location | 9,405,032 – 9,405,134 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -25.88 |
| Energy contribution | -24.77 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
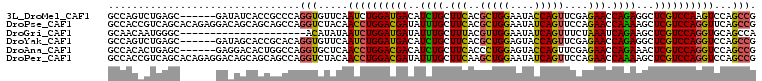
>3L_DroMel_CAF1 9405032 102 + 23771897 GCCAGUCUGAGC------GAUAUCACCGCCCAGGUGUUCAAUCUGGAUGACAUCUGCUUCACGCUGGAAUACCAGUUCGAGAACCAGAGGCUCGUCCAAGUCCAGCCG ....(.(((..(------((...((((.....)))).))....(((((((..((((.(((.((((((....))))..)).))).))))...))))))).)..))).). ( -33.40) >DroPse_CAF1 1891 108 + 1 GCCACCGUCAGCACAGAGGACAGCAGCAGCCAGGUCUACAACCUGGACGAUAUUUGCUUCACGCUGGAAUAUCAGUUCCAGAACCAAAAGCUCGUCCAGGUUCAGCCG ..........((...(.((((.((....))...)))).)(((((((((((.....((((....(((((((....)))))))......)))))))))))))))..)).. ( -36.00) >DroGri_CAF1 1858 87 + 1 GCAACAAUGGGC---------------------ACAUAUAAUCUGGAUGAUAUUUGCUUUACGUUGGAAUAUCAGUUUCUAAAUCAGAAGCUCGUCCAGGUGCAGCCA .........(((---------------------.......((((((((((.....(((((...((((((.......))))))....)))))))))))))))...))). ( -25.00) >DroYak_CAF1 866 102 + 1 GCCAGUCUGAGC------GAUAGCACCGCACAGGUGUUCAAUCUGGAUGACAUCUGCUUCACGCUGGAGUACCAGUUCGAGAACCAGAGGCUCGUCCAGGUCCAGCCG ....(.(((..(------((..(((((.....)))))))...((((((((..((((.(((.((((((....))))..)).))).))))...)))))))))..))).). ( -37.00) >DroAna_CAF1 1825 102 + 1 GCCACACUGAGC------GAGGACACUGGCCAGGUGCUCAACCUGGACGACAUCUGCUUCACCCUGGAGUACCAGUUCGAGAACCAGAAACUCGUCCAGGUCCAGCCG ...((.(((.((------.((....)).)))))))(((..((((((((((..((((.(((.(.((((....))))...).))).))))...))))))))))..))).. ( -40.60) >DroPer_CAF1 8475 108 + 1 GCCACCGUCAGCACAGAGGACAGCAGCAGCCAGGUCUACAACCUGGACGAUAUUUGCUUCAAGCUGGAAUAUCAGUUCCAGAACCAAAAGCUCGUCCAGGUCCAGCCG ..........((...(.((((.((....))...)))).).((((((((((.....((((....(((((((....)))))))......))))))))))))))...)).. ( -35.10) >consensus GCCACACUGAGC______GACAGCACCGGCCAGGUGUACAACCUGGACGACAUCUGCUUCACGCUGGAAUACCAGUUCCAGAACCAGAAGCUCGUCCAGGUCCAGCCG ................................(((.....((((((((((..((((.(((..(((((....)))))....))).))))...))))))))))...))). (-25.88 = -24.77 + -1.11)
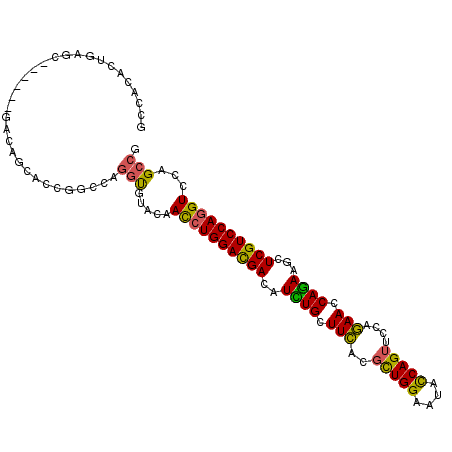
| Location | 9,405,054 – 9,405,169 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.66 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -25.08 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9405054 115 + 23771897 CCCAGGUGUUCAAUCUGGAUGACAUCUGCUUCACGCUGGAAUACCAGUUCGAGAACCAGAGGCUCGUCCAAGUCCAGCCGCCCACUGUGGUCUCGCUCAGCAUGGUGUCCAGCCC----- .((((((.....))))))................((((((.(((((((((((((.((((.((((.(.......).)))).....))).).)))))...))).)))))))))))..----- ( -38.20) >DroPse_CAF1 1919 115 + 1 GCCAGGUCUACAACCUGGACGAUAUUUGCUUCACGCUGGAAUAUCAGUUCCAGAACCAAAAGCUCGUCCAGGUUCAGCCGCCAACUGUGGUGUCCUUGAGCAUGCUAACCACAUC----- ((((((.....(((((((((((.....((((....(((((((....)))))))......)))))))))))))))..(((((.....)))))...)))).))..............----- ( -40.20) >DroGri_CAF1 1870 107 + 1 -----ACAUAUAAUCUGGAUGAUAUUUGCUUUACGUUGGAAUAUCAGUUUCUAAAUCAGAAGCUCGUCCAGGUGCAGCCACCCACAGUGGUGUCGUUCAGCAUGUUAUCGCC-------- -----....((((((((((((((((..(((.(((.(((((.....(((((((.....)))))))..)))))))).)))(((.....))))))))))))))...)))))....-------- ( -29.30) >DroWil_CAF1 772 120 + 1 AACAGGUUUAUAAUUUGGAUGACAUAUGCUUCACCUUGGAAUAUCAAUUUCAAAAUCAAAAACUUGUCCAAGUCCAGCCACCCACAGUGGUAUCGCUCAGCAUGCUGGCCGCCUCAUCCU ...((((........(((((((((...........(((((((....)))))))...........))))...)))))(((((.....)))))...((.(((....))))).))))...... ( -25.05) >DroAna_CAF1 1847 115 + 1 GCCAGGUGCUCAACCUGGACGACAUCUGCUUCACCCUGGAGUACCAGUUCGAGAACCAGAAACUCGUCCAGGUCCAGCCGCCCACCGUCGUCUCGCUUAGCAUGAUGGCCCCCGC----- ....((((((..((((((((((..((((.(((.(.((((....))))...).))).))))...))))))))))..)).))))..(((((((((.....)).))))))).......----- ( -42.30) >DroPer_CAF1 8503 115 + 1 GCCAGGUCUACAACCUGGACGAUAUUUGCUUCAAGCUGGAAUAUCAGUUCCAGAACCAAAAGCUCGUCCAGGUCCAGCCGCCAACUGUGGUGUCCUUGAGCAUGCUAACCACAUC----- ((((((......((((((((((.....((((....(((((((....)))))))......))))))))))))))...(((((.....)))))...)))).))..............----- ( -39.30) >consensus GCCAGGUCUACAACCUGGACGACAUUUGCUUCACGCUGGAAUAUCAGUUCCAGAACCAAAAGCUCGUCCAGGUCCAGCCGCCCACUGUGGUGUCGCUCAGCAUGCUAACCACCUC_____ ....(((.....((((((((((..((((.(((..(((((....)))))....))).))))...))))))))))...(((((.....)))))................))).......... (-25.08 = -24.42 + -0.66)

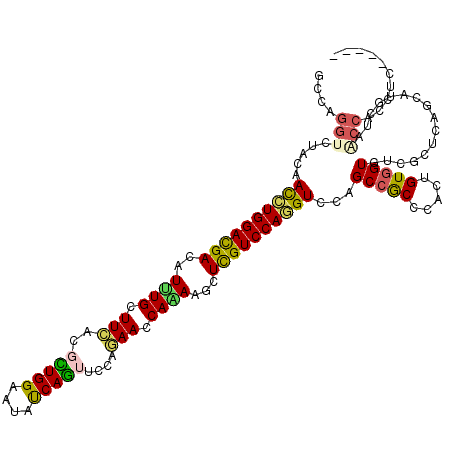
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:26 2006