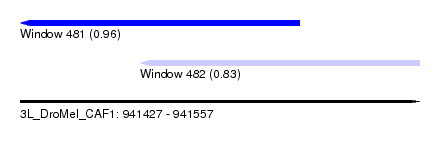
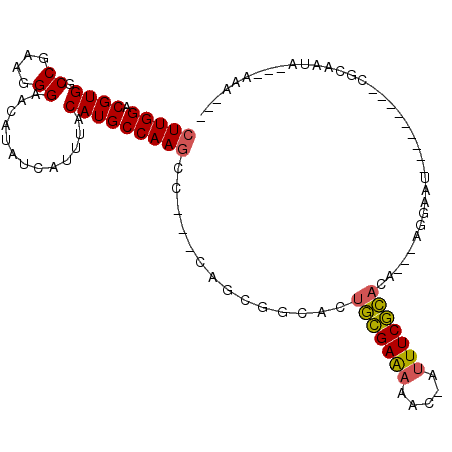
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 941,427 – 941,557 |
| Length | 130 |
| Max. P | 0.960666 |

| Location | 941,427 – 941,518 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -12.73 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 941427 91 - 23771897 AGCCAGCCAGCGGCACUGCGAAAAAUCUUUCGCACA---AGGAAU---------CGCAAUAAAA----AUUCUAAUUUGCAUACAUUUUUUGCUUCCAAGAACUCGU .(((.......)))..(((((((....)))))))..---.((((.---------.((((.....----........))))...((.....)).)))).......... ( -16.62) >DroSec_CAF1 75189 91 - 1 AGCCAGCCAGCGGCACUGCGAAAAACAUUUCGCACA---AGGAAU---------CGCAAUAAAA----AAUCUAAUUUGCAUAAAUUUAUUGCUUCCAAAAACUGGU .....(((((......(((((((....)))))))..---.((((.---------.((((((((.----.................)))))))))))).....))))) ( -21.97) >DroSim_CAF1 68389 91 - 1 AGCCAGCCAGCGGCACUGCGAAAAACAUUUCGCACA---AGGAAU---------CGCAAUAAAA----AUUCUAAUUUGCAUAAAUUUAUUGCUUCCAAAAACUGGU .....(((((......(((((((....)))))))..---.((((.---------.((((((((.----.................)))))))))))).....))))) ( -21.97) >DroYak_CAF1 79171 100 - 1 AGC----CAGCGGCACUGCGAGAAAUAUUUCGCACAUGCAGCAAGCAAAAACUUCACAAUGAAAUAGAAUUGAGAUUUACAGAUGUUUAUUGGUCUCAAGAACU--- .((----..((.(((.(((((((....)))))))..))).))..)).......................((((((((.((....)).....)))))))).....--- ( -22.60) >consensus AGCCAGCCAGCGGCACUGCGAAAAACAUUUCGCACA___AGGAAU_________CGCAAUAAAA____AUUCUAAUUUGCAUAAAUUUAUUGCUUCCAAAAACUGGU .(((.......)))..(((((((....))))))).....................((((((((......................)))))))).............. (-12.73 = -13.10 + 0.37)

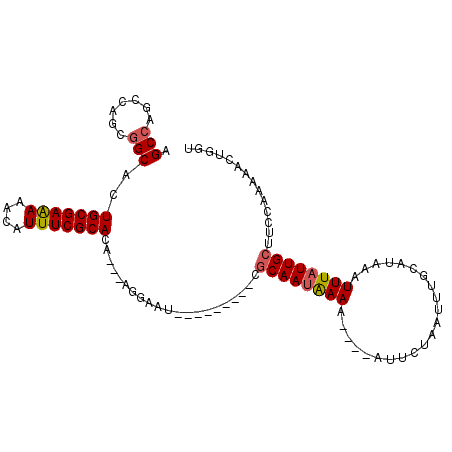
| Location | 941,466 – 941,557 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -16.63 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 941466 91 - 23771897 CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGCCAGCCAGCGGCACUGCGAAAAAU-CUUUCGCACA---AGGAAU---------CGCAAUA---AAA--- (((((.((((.((....)).............)))))))))((.(((...)))..(((((((...-.)))))))..---.))...---------.......---...--- ( -23.34) >DroSec_CAF1 75228 91 - 1 CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGCCAGCCAGCGGCACUGCGAAAAAC-AUUUCGCACA---AGGAAU---------CGCAAUA---AAA--- (((((.((((.((....)).............)))))))))((.(((...)))..(((((((...-.)))))))..---.))...---------.......---...--- ( -23.14) >DroSim_CAF1 68428 91 - 1 CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGCCAGCCAGCGGCACUGCGAAAAAC-AUUUCGCACA---AGGAAU---------CGCAAUA---AAA--- (((((.((((.((....)).............)))))))))((.(((...)))..(((((((...-.)))))))..---.))...---------.......---...--- ( -23.14) >DroEre_CAF1 78367 92 - 1 CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGC----CAGCGGCACUGCGAGAAAC-AUUUCGCACUUGCAGGAAUCAAAAACUCAAG------------- (((((.((((.((....)).............)))))))))(----(.((((...(((((((...-.))))))).)))).))...............------------- ( -23.04) >DroYak_CAF1 79208 102 - 1 CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGC----CAGCGGCACUGCGAGAAAU-AUUUCGCACAUGCAGCAAGCAAAAACUUCACAAUG---AAAUAG .((((.((((.((....)).............))))))))((----..((.(((.(((((((...-.)))))))..))).))..))...............---...... ( -25.14) >DroAna_CAF1 82857 85 - 1 GUUGGACGUGGCCGA-GGGAACAUAUCAUUUACAUGCCAAGUU----AGAUACACCAAGAGAAAUCAAUUCUU--------GAAA---------CUUUAUAUAUAAA--- ((..(((.((((...-.(((........)))....)))).)))----....))..((((((.......)))))--------)...---------.............--- ( -10.50) >consensus CUUGGACGUGGCCGAAGGGAACAUAUCAUUUACAUGCCAAGCC___CAGCGGCACUGCGAAAAAC_AUUUCGCACA___AGGAAU_________CGCAAUA___AAA___ (((((.((((.((....)).............)))))))))..............(((((((.....))))))).................................... (-16.63 = -16.32 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:12 2006