
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,397,565 – 9,397,670 |
| Length | 105 |
| Max. P | 0.797927 |

| Location | 9,397,565 – 9,397,670 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -33.77 |
| Energy contribution | -33.38 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9397565 105 + 23771897 CAGCUGAAUUGCAAUGGCAAUGGCACCACCGGUGGCGCGGCGAAAACCACUGUGGCCUCGGCCAUAACACCGCCCAAGCAGCCCCAGUUGGCGGCCAAGGUCAUC ..((......)).(((((..((((.((((.((.(((((((((........((((((....))))))....))))...)).))))).).)))..))))..))))). ( -39.10) >DroSec_CAF1 20305 105 + 1 CAGCUGAAUUGCAAUGGCAAUGGCACCACCGAUGGCGCGGCCAAAACCACUGUGGCCUCGGCCAUAACACCGCCCAAGCAGCCCCAGCUGGCGGCCAAGGUCAUC ((((((..((((...(((..((((..(.((...)).)..)))).......((((((....)))))).....)))...))))...))))))..(((....)))... ( -38.50) >DroSim_CAF1 19845 105 + 1 CAGCUGAAUUGCAAUGGCAAUGGCACCACCGGUGGCGCGGCCAAAACCACUGUGGCCUCGGCCAUAACACCGCCCAAGCAGCCCCAGCUGGCGGCCAAGGUCAUC ((((((..((((..((((..((.((((...)))).))..)))).......((((((....))))))...........))))...))))))..(((....)))... ( -39.70) >DroEre_CAF1 21612 105 + 1 CAGCUGAAUUGCAAUGGCAAUGGCACCACCGGUGGGGCUGCCAAAACCACUGUGGCCUCGGCCAUAACACCGCCCAAGCAGCCCCAAUUGGCGGCCAAGGUCAUC ..((......)).(((((..((((....(((((((((((((.........((((((....))))))...........)))))))).)))))..))))..))))). ( -44.25) >DroYak_CAF1 18399 105 + 1 CAGCUGAAUUGCAAUGGCAAUGGCACCACCGGUGGGGCGGCCAAAACCACUGUGGCCUCGGCCAUAACGCCGCCCAAGCAGCCCCAAUUGGCGGCCAAGGUCAUC ..((......)).(((((..((((....(((((((((((((((.........))))).((((......))))........))))).)))))..))))..))))). ( -43.00) >DroAna_CAF1 21434 105 + 1 CAGCUGAACUGUAAUGGUAAUGGCACUUCGGGAGGAGCUGCCAAGACCACUGUGGCCUCGGCCAUAACACCGCCGAAACAACCCCAGUUGGCGGCCAAGGUCAUC ..(((((.(..((.((((..(((((((((....)))).)))))..)))).))..)..)))))......(((((((...((((....)))).))))...))).... ( -39.80) >consensus CAGCUGAAUUGCAAUGGCAAUGGCACCACCGGUGGCGCGGCCAAAACCACUGUGGCCUCGGCCAUAACACCGCCCAAGCAGCCCCAGUUGGCGGCCAAGGUCAUC ((((((..((((...(((..((((.(((....)))....)))).......((((((....)))))).....)))...))))...))))))..(((....)))... (-33.77 = -33.38 + -0.39)

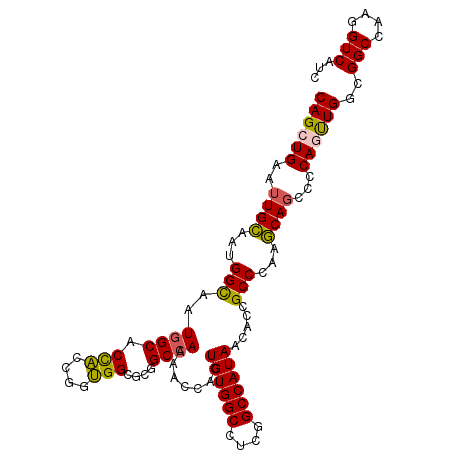

| Location | 9,397,565 – 9,397,670 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -40.10 |
| Energy contribution | -39.77 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9397565 105 - 23771897 GAUGACCUUGGCCGCCAACUGGGGCUGCUUGGGCGGUGUUAUGGCCGAGGCCACAGUGGUUUUCGCCGCGCCACCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ..(((....((((.((....))))))((...(((((((....(((.((((((.....)))))).)))((((((....)))))))))))))...))...))).... ( -43.60) >DroSec_CAF1 20305 105 - 1 GAUGACCUUGGCCGCCAGCUGGGGCUGCUUGGGCGGUGUUAUGGCCGAGGCCACAGUGGUUUUGGCCGCGCCAUCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ...............((((((((..(((...(((((((....((((((((((.....))))))))))((((((....)))))))))))))...))).)))))))) ( -52.00) >DroSim_CAF1 19845 105 - 1 GAUGACCUUGGCCGCCAGCUGGGGCUGCUUGGGCGGUGUUAUGGCCGAGGCCACAGUGGUUUUGGCCGCGCCACCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ...............((((((((..(((...(((((((....((((((((((.....))))))))))((((((....)))))))))))))...))).)))))))) ( -52.90) >DroEre_CAF1 21612 105 - 1 GAUGACCUUGGCCGCCAAUUGGGGCUGCUUGGGCGGUGUUAUGGCCGAGGCCACAGUGGUUUUGGCAGCCCCACCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ..((.(((((((((((....)).(((((....)))))....))))))))).))(((((((..(((((.(((....)).).)))))..)))))))........... ( -46.50) >DroYak_CAF1 18399 105 - 1 GAUGACCUUGGCCGCCAAUUGGGGCUGCUUGGGCGGCGUUAUGGCCGAGGCCACAGUGGUUUUGGCCGCCCCACCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ..((.(((((((((((....)).(((((....)))))....))))))))).))(((((((..((((.(((((...)).)))))))..)))))))........... ( -47.80) >DroAna_CAF1 21434 105 - 1 GAUGACCUUGGCCGCCAACUGGGGUUGUUUCGGCGGUGUUAUGGCCGAGGCCACAGUGGUCUUGGCAGCUCCUCCCGAAGUGCCAUUACCAUUACAGUUCAGCUG ((((....(((((..((((....)))).(((((.((.((....(((((((((.....))))))))).)).))..)))))).))))....)))).(((.....))) ( -37.40) >consensus GAUGACCUUGGCCGCCAACUGGGGCUGCUUGGGCGGUGUUAUGGCCGAGGCCACAGUGGUUUUGGCCGCGCCACCGGUGGUGCCAUUGCCAUUGCAAUUCAGCUG ..((.(((((((((((....)).(((((....)))))....))))))))).))(((((((..((((....(((....))).))))..)))))))........... (-40.10 = -39.77 + -0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:22 2006