
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,389,891 – 9,390,131 |
| Length | 240 |
| Max. P | 0.994281 |

| Location | 9,389,891 – 9,389,995 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -23.83 |
| Energy contribution | -24.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9389891 104 + 23771897 AGAUUUAAUCA-UUUACUGGACAGGUAAACCGGCAUCGCCGUGUCCAAGUUCCGGGGACCGUACUAAAUAGAUCGUGGUCUGGCAGUUGGGGCAUGUGGUGCACA ...........-((((((.....))))))...(((((((.((((((.(((.(.(....).).)))...(((((....))))).......)))))))))))))... ( -29.10) >DroEre_CAF1 13174 104 + 1 AGAUUUCAUCA-UUUACUGGACAGGUAAACCGGCAUCGCCAUGUCCAAGUUCCGGGGAGCGUACUAAAUAUAUCGUGGUCUGGCAGUUGGGGCAUGUGGUGCACA ...........-((((((.....))))))...(((((((.((((((((.(.(((((..(((............)))..))))).).)).)))))))))))))... ( -31.20) >DroWil_CAF1 19869 95 + 1 ---------GG-CUUAUUGCACUGGGGAACCAGCAUCACCGUGUCCCAGCUCGGGGGAGCGUACCAAAUAGAUGGUUGUUUGGCAAUUGGGACAAGUGGUGCACA ---------.(-(.....))..(((....)))(((((((..(((((((((((....))))...((((((((....))))))))....))))))).)))))))... ( -40.70) >DroYak_CAF1 10655 103 + 1 AGAUUU-AUCG-UUUACUGGACUGGUAAGCCGGCAUCGCCAUGUCCCAAUUCCGGGGAGCGUACUAAAUAGAUCGUGGUCUGGCAGUUGGGGCAUGUGGUGCACA ......-...(-((((((.....)))))))..(((((((.((((((((((((((((..(((..((....))..)))..))))).))))))))))))))))))... ( -43.20) >DroMoj_CAF1 14821 99 + 1 ----UUGGUCAU--UAUUGCAUUGGCAUACCAGCAUCGCCAUGGCCCAGCUCAGGUGUGCGCACCAGGUAAAUGGUUGUCUGGCAAUUGGGGCAUGUGGUGCACA ----.((((...--...(((....))).))))(((((((.(((.((((((.((....)).)).((((..((....))..))))....)))).))))))))))... ( -34.60) >DroAna_CAF1 13373 98 + 1 GGGUU-------UCUACUGUACUGGUGACCCGGCAUCUCCGUGUCCCAGUUCGGGGGAGCGUACCAAGUAAAUAGUGGUCUGGCAGUUGGGGCACGUGGUGCACA (((((-------.(((......))).))))).(((((..(((((((((((((....))))((((((.........))))...))...))))))))).)))))... ( -37.50) >consensus AGAUUU_AUCA_UUUACUGGACUGGUAAACCGGCAUCGCCAUGUCCCAGUUCCGGGGAGCGUACCAAAUAGAUCGUGGUCUGGCAGUUGGGGCAUGUGGUGCACA .......................((....)).(((((((.((((((((((((....))))...(((.((........)).)))....)))))))))))))))... (-23.83 = -24.03 + 0.20)


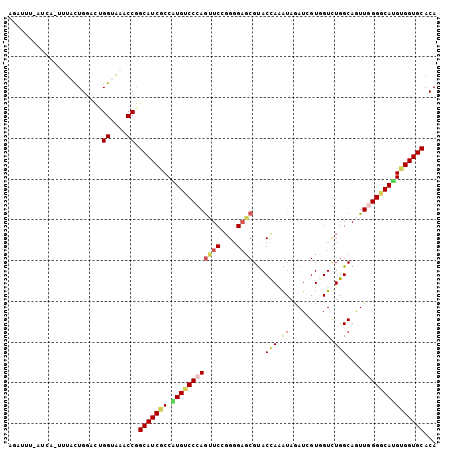
| Location | 9,389,995 – 9,390,104 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9389995 109 + 23771897 UCUCGGCUUCCGCCGGUGCAGCAGAGCCACCAGCGUUUUCUCUGGGUGCAUUACUGCCGCCAGUUGCUCCUGGAGCUGGUGCUGCU---CCUCCUCCGGUGGUUGGCAUAGU .....(((.(((((((.(.((..(((((((((((......((..((.(((..((((....))))))).))..)))))))))..)))---))).).)))))))..)))..... ( -48.70) >DroEre_CAF1 13278 112 + 1 UCUCGGCAUCCGCCGGUGCCGCAGAGCCACCAGCGUUUUCCCUGGGCGCAUUACUGCCGCCAGUUGCUCCUGGAGCUGCUGGUGCUGCUGCUCCUCCGGUGGUUGGCAUAGA ....(.((.(((((((.(..(((((((.(((((((..((((..(((.(((..((((....))))))).))))))).)))))))))).))))..).))))))).)).)..... ( -52.90) >DroWil_CAF1 19964 87 + 1 UUUCAGCAUCCGGUGGGGGAGCAGCACCACCAGCUC------CG---CCAUUAUUGCC------------AGAAGCAGCCGCUGCCGCAGCAUUU----UGAUUAGCAUUGG .....(((...(((((.(((((..........))))------).---)))))..)))(------------((((...((.((....)).)).)))----))........... ( -24.90) >DroYak_CAF1 10758 109 + 1 UCUCGGCAUCCGCCGGUGCCGCAGAGCCACCAGCAUUUUCCCUGGGCGCAUUAUUGCCGCCAGUUGCUCCAGGAGCUGCG---GCUGCUCCUCCUCCGGUGGUUGGUGUAGA (((((.((.(((((((.(((((((..((...((((......(((((.(((....)))).)))).))))...))..)))))---))..........))))))).)).)).))) ( -47.10) >consensus UCUCGGCAUCCGCCGGUGCAGCAGAGCCACCAGCGUUUUCCCUGGGCGCAUUACUGCCGCCAGUUGCUCCAGGAGCUGCUGCUGCUGCUCCUCCUCCGGUGGUUGGCAUAGA ..(((((....)))))....((...((((((.............((((((....)).))))..........(((((.((.......)).)))))...))))))..))..... (-22.50 = -23.75 + 1.25)



| Location | 9,390,032 – 9,390,131 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.52 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -17.34 |
| Energy contribution | -19.39 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9390032 99 - 23771897 CCGCCGAAUGCGGUUGCUCCGCAGGCAACUAU---GCCAACCACCGGAGGAGG---AGCAGCACCAGCUCCAGGAGCAACUGGCGGCAGUAAUGCACCCAGAGAA ((((.....))))(((((((...((((....)---)))..((......)).((---(((.......))))).)))))))((((..(((....)))..)))).... ( -43.50) >DroSec_CAF1 12932 102 - 1 CCGCCGAAUGCGGUUGCUCCGCAGGCAACUAU---GCCAACCACCGGAGGAGCAGGAGCAGCACCAGCUCCAGGAGCAACUGGCGGCAGUAAUGCACCCAGAGAA ((((.....))))(((((((((.((((....)---)))..((......)).)).(((((.......))))).)))))))((((..(((....)))..)))).... ( -46.60) >DroSim_CAF1 12430 102 - 1 CCGCCGAAUGCGGUUGCUCCGCAGGCAACUAU---GCCAACCACCGGAGGAGGAGCAGCAGCACAAGCUCCAGGAGCAACUGGCGGCAGUAAUGCACCCAGAGAA ((((.....))))(((((((...((((....)---)))..((......)).(((((..........))))).)))))))((((..(((....)))..)))).... ( -42.90) >DroEre_CAF1 13315 96 - 1 CCGCCAAAU------GCUCCGCAGGCAUCUAU---GCCAACCACCGGAGGAGCAGCAGCACCAGCAGCUCCAGGAGCAACUGGCGGCAGUAAUGCGCCCAGGGAA .(((((..(------(((((...((((....)---)))..........(((((.((.......)).))))).))))))..)))))(((....))).......... ( -41.40) >DroYak_CAF1 10795 93 - 1 CCUCCAAAC------GCUCCACAGACAUCUAC---ACCAACCACCGGAGGAGGAGCAGC---CGCAGCUCCUGGAGCAACUGGCGGCAAUAAUGCGCCCAGGGAA ..(((....------(((((............---.....((......))((((((...---....)))))))))))..((((.((((....))).)))))))). ( -32.50) >DroAna_CAF1 13508 94 - 1 -GGCCGGAGGAAGU-------CAGGCAGCUGGAGCACCAUCAAGUGGCGGACUAGGAGCCGGU---GCAGCAGGAGGAGCCGGCAAUGCCGCUGCACCACGAGAG -(((........))-------).(((..((((..(.((((...)))).)..))))..)))(((---(((((.((.....))(((...)))))))))))....... ( -35.70) >consensus CCGCCGAAUGCGGU_GCUCCGCAGGCAACUAU___GCCAACCACCGGAGGAGGAGCAGCAGCACCAGCUCCAGGAGCAACUGGCGGCAGUAAUGCACCCAGAGAA ...............(((((...(((.........)))..........((((...............)))).)))))..((((..(((....)))..)))).... (-17.34 = -19.39 + 2.06)


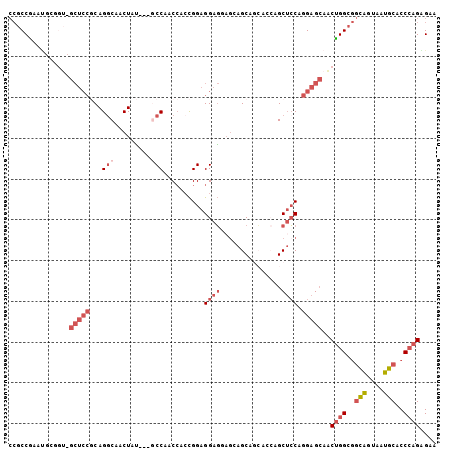
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:19 2006